MicroChIP--a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies
- PMID: 18202078
- PMCID: PMC2241883
- DOI: 10.1093/nar/gkm1158
MicroChIP--a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies
Abstract
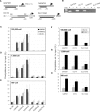
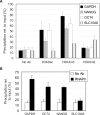
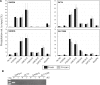
Chromatin immunoprecipitation (ChIP) is a powerful technique for studying protein-DNA interactions. Drawbacks of current ChIP assays however are a requirement for large cell numbers, which limits applicability of ChIP to rare cell samples, and/or lengthy procedures with limited applications. There are to date no protocols for fast and parallel ChIPs of post-translationally modified histones from small cell numbers or biopsies, and importantly, no protocol allowing for investigations of transcription factor binding in small cell numbers. We report here the development of a micro (micro) ChIP assay suitable for up to nine parallel quantitative ChIPs of modified histones or RNA polymerase II from a single batch of 1000 cells. MicroChIP can also be downscaled to monitor the association of one protein with multiple genomic sites in as few as 100 cells. MicroChIP is applicable to small fresh tissue biopsies, and a cross-link-while-thawing procedure makes the assay suitable for frozen biopsies. Using MicroChIP, we characterize transcriptionally permissive and repressive histone H3 modifications on developmentally regulated promoters in human embryonal carcinoma cells and in osteosarcoma biopsies. muChIP creates possibilities for multiple parallel and rapid transcription factor binding and epigenetic analyses of rare cell and tissue samples.
Figures
Similar articles
-
A rapid micro chromatin immunoprecipitation assay (microChIP).Nat Protoc. 2008;3(6):1032-45. doi: 10.1038/nprot.2008.68. Nat Protoc. 2008. PMID: 18536650
-
MicroChIP: chromatin immunoprecipitation for small cell numbers.Methods Mol Biol. 2009;567:59-74. doi: 10.1007/978-1-60327-414-2_4. Methods Mol Biol. 2009. PMID: 19588085 Review.
-
Micro chromatin immunoprecipitation (μChIP) from early mammalian embryos.Methods Mol Biol. 2015;1222:227-45. doi: 10.1007/978-1-4939-1594-1_17. Methods Mol Biol. 2015. PMID: 25287350
-
A quick and quantitative chromatin immunoprecipitation assay for small cell samples.Front Biosci. 2007 Sep 1;12:4925-31. doi: 10.2741/2438. Front Biosci. 2007. PMID: 17569620
-
Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond.Cell Cycle. 2014;13(18):2847-52. doi: 10.4161/15384101.2014.949201. Cell Cycle. 2014. PMID: 25486472 Free PMC article. Review.
Cited by
-
DNA methylation directs functional maturation of pancreatic β cells.J Clin Invest. 2015 Jul 1;125(7):2851-60. doi: 10.1172/JCI79956. Epub 2015 Jun 22. J Clin Invest. 2015. PMID: 26098213 Free PMC article.
-
Combining genomic and proteomic approaches for epigenetics research.Epigenomics. 2013 Aug;5(4):439-52. doi: 10.2217/epi.13.37. Epigenomics. 2013. PMID: 23895656 Free PMC article. Review.
-
Limitations and possibilities of low cell number ChIP-seq.BMC Genomics. 2012 Nov 21;13:645. doi: 10.1186/1471-2164-13-645. BMC Genomics. 2012. PMID: 23171294 Free PMC article.
-
Integrated genome-wide analysis of transcription factor occupancy, RNA polymerase II binding and steady-state RNA levels identify differentially regulated functional gene classes.Nucleic Acids Res. 2012 Jan;40(1):148-58. doi: 10.1093/nar/gkr720. Epub 2011 Sep 13. Nucleic Acids Res. 2012. PMID: 21914722 Free PMC article.
-
Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation.Proc Natl Acad Sci U S A. 2010 Aug 24;107(34):15081-6. doi: 10.1073/pnas.1009575107. Epub 2010 Aug 9. Proc Natl Acad Sci U S A. 2010. PMID: 20696891 Free PMC article.
References
-
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. - PubMed
-
- O’Neill LP, Turner BM. Immunoprecipitation of chromatin. Methods Enzymol. 1996;274:189–197. - PubMed
-
- O’Neill LP, Vermilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat. Genet. 2006;38:835–841. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

