The foldon substructure of staphylococcal nuclease
- PMID: 18201720
- PMCID: PMC2268249
- DOI: 10.1016/j.jmb.2007.12.020
The foldon substructure of staphylococcal nuclease
Abstract
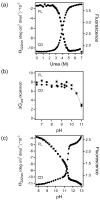
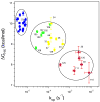
To search for submolecular foldon units, the spontaneous reversible unfolding and refolding of staphylococcal nuclease under native conditions was studied by a kinetic native-state hydrogen exchange (HX) method. As for other proteins, it appears that staphylococcal nuclease is designed as an assembly of well-integrated foldon units that may define steps in its folding pathway and may regulate some other functional properties. The HX results identify 34 amide hydrogens that exchange with solvent hydrogens under native conditions by way of large transient unfolding reactions. The HX data for each hydrogen measure the equilibrium stability (Delta G(HX)) and the kinetic unfolding and refolding rates (k(op) and k(cl)) of the unfolding reaction that exposes it to exchange. These parameters separate the 34 identified residues into three distinct HX groupings. Two correspond to clearly defined structural units in the native protein, termed the blue and red foldons. The remaining HX grouping contains residues, not well separated by their HX parameters alone, that represent two other distinct structural units in the native protein, termed the green and yellow foldons. Among these four sets, a last unfolding foldon (blue) unfolds with a rate constant of 6 x 10(-6) s(-1) and free energy equal to the protein's global stability (10.0 kcal/mol). It represents part of the beta-barrel, including mutually H-bonding residues in the beta 4 and beta 5 strands, a part of the beta 3 strand that H-bonds to beta 5, and residues at the N-terminus of the alpha2 helix that is capped by beta 5. A second foldon (green), which unfolds and refolds more rapidly and at slightly lower free energy, includes residues that define the rest of the native alpha2 helix and its C-terminal cap. A third foldon (yellow) defines the mutually H-bonded beta1-beta2-beta 3 meander, completing the native beta-barrel, plus an adjacent part of the alpha1 helix. A final foldon (red) includes residues on remaining segments that are distant in sequence but nearly adjacent in the native protein. Although the structure of the partially unfolded forms closely mimics the native organization, four residues indicate the presence of some nonnative misfolding interactions. Because the unfolding parameters of many other residues are not determined, it seems likely that the concerted foldon units are more extensive than is shown by the 34 residues actually observed.
Figures


Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Unlocking data: Decision-maker perspectives on cross-sectoral data sharing and linkage as part of a whole-systems approach to public health policy and practice.Public Health Res (Southampt). 2024 Nov 20:1-30. doi: 10.3310/KYTW2173. Online ahead of print. Public Health Res (Southampt). 2024. PMID: 39582242
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
High-Resolution Mapping of a Repeat Protein Folding Free Energy Landscape.Biophys J. 2016 Dec 6;111(11):2368-2376. doi: 10.1016/j.bpj.2016.08.027. Biophys J. 2016. PMID: 27926838 Free PMC article.
-
NMR Analysis of Amide Hydrogen Exchange Rates in a Pentapeptide-Repeat Protein from A. thaliana.Biophys J. 2017 May 23;112(10):2075-2088. doi: 10.1016/j.bpj.2017.04.016. Biophys J. 2017. PMID: 28538145 Free PMC article.
-
Conditionally and transiently disordered proteins: awakening cryptic disorder to regulate protein function.Chem Rev. 2014 Jul 9;114(13):6779-805. doi: 10.1021/cr400459c. Epub 2014 Feb 6. Chem Rev. 2014. PMID: 24502763 Free PMC article. Review. No abstract available.
-
The case for defined protein folding pathways.Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):8253-8258. doi: 10.1073/pnas.1706196114. Epub 2017 Jun 19. Proc Natl Acad Sci U S A. 2017. PMID: 28630329 Free PMC article.
-
Simplified protein models: predicting folding pathways and structure using amino acid sequences.Phys Rev Lett. 2013 Jul 12;111(2):028103. doi: 10.1103/PhysRevLett.111.028103. Epub 2013 Jul 11. Phys Rev Lett. 2013. PMID: 23889448 Free PMC article.
References
-
- Bai Y, Englander SW. Future directions in folding: The multi-state nature of protein structure. Proteins: Struct Funct Genet. 1996;24:145–51. - PubMed
-
- Englander SW. Protein folding intermediates and pathways studied by hydrogen exchange. Annu Rev Biophys Biomol Struct. 2000;29:213–238. - PubMed
-
- Krishna MMG, Lin Y, Mayne L, Englander SW. Intimate view of a kinetic protein folding intermediate: Residue-resolved structure, interactions, stability, folding and unfolding rates, homogeneity. J Mol Biol. 2003;334:501–513. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

