Complement evasion by human pathogens
- PMID: 18197169
- PMCID: PMC2814840
- DOI: 10.1038/nrmicro1824
Complement evasion by human pathogens
Abstract
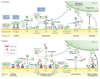
The human immune system has developed an elaborate network of cascades for dealing with microbial intruders. Owing to its ability to rapidly recognize and eliminate microorganisms, the complement system is an essential and efficient component of this machinery. However, many pathogenic organisms have found ways to escape the attack of complement through a range of different mechanisms. Recent discoveries in this field have provided important insights into these processes on a molecular level. These vital developments could augment our knowledge of the pathology and treatment of infectious and inflammatory diseases.
Figures

Similar articles
-
Complement evasion strategies of microorganisms.Immunol Today. 1991 Sep;12(9):327-31. doi: 10.1016/0167-5699(91)90010-Q. Immunol Today. 1991. PMID: 1721820 Review.
-
Viral mimicry of the complement system.J Biosci. 2003 Apr;28(3):249-64. doi: 10.1007/BF02970145. J Biosci. 2003. PMID: 12734404 Free PMC article. Review.
-
Viral regulators of complement activation: structure, function and evolution.Mol Immunol. 2014 Oct;61(2):89-99. doi: 10.1016/j.molimm.2014.06.004. Epub 2014 Jun 27. Mol Immunol. 2014. PMID: 24976595 Review.
-
[The efficiency of the bactericidal action of serum raised by complement and lysozyme against bacteria which avoid the immunological response of higher organisms].Postepy Hig Med Dosw (Online). 2009 Oct 19;63:471-84. Postepy Hig Med Dosw (Online). 2009. PMID: 19850971 Review. Polish.
-
Complement inhibition by gram-positive pathogens: molecular mechanisms and therapeutic implications.J Mol Med (Berl). 2010 Feb;88(2):115-20. doi: 10.1007/s00109-009-0572-y. J Mol Med (Berl). 2010. PMID: 20062962 Free PMC article. Review.
Cited by
-
Innate immune response to oral bacteria and the immune evasive characteristics of periodontal pathogens.J Periodontal Implant Sci. 2013 Feb;43(1):3-11. doi: 10.5051/jpis.2013.43.1.3. Epub 2013 Feb 28. J Periodontal Implant Sci. 2013. PMID: 23507986 Free PMC article.
-
Moraxella catarrhalis Binds Plasminogen To Evade Host Innate Immunity.Infect Immun. 2015 Sep;83(9):3458-69. doi: 10.1128/IAI.00310-15. Epub 2015 Jun 22. Infect Immun. 2015. PMID: 26099590 Free PMC article.
-
Natural mutations in a Staphylococcus aureus virulence regulator attenuate cytotoxicity but permit bacteremia and abscess formation.Proc Natl Acad Sci U S A. 2016 May 31;113(22):E3101-10. doi: 10.1073/pnas.1520255113. Epub 2016 May 16. Proc Natl Acad Sci U S A. 2016. PMID: 27185949 Free PMC article.
-
Role of factor H binding protein in Neisseria meningitidis virulence and its potential as a vaccine candidate to broadly protect against meningococcal disease.Microbiol Mol Biol Rev. 2013 Jun;77(2):234-52. doi: 10.1128/MMBR.00056-12. Microbiol Mol Biol Rev. 2013. PMID: 23699256 Free PMC article. Review.
-
The complement system.Cell Tissue Res. 2011 Jan;343(1):227-35. doi: 10.1007/s00441-010-1034-0. Epub 2010 Sep 14. Cell Tissue Res. 2011. PMID: 20838815 Free PMC article. Review.
References
-
- Lambris JD, Sahu A, Wetsel RA. In: The Human Complement System in Health and Disease. Volanakis JE, Frank MM, editors. New York: Marcel Dekker; 1998. pp. 83–118.
-
- Lambris JD, Holers VM, editors. Therapeutic Interventions in the Complement System. Totowa: Humana; 2000.
-
- Walport MJ. Complement. First of two parts. N. Engl. J. Med. 2001;344:1058–1066. - PubMed
-
-
Walport MJ. Complement. Second of two parts. N. Engl. J. Med. 2001;344:1140–1144. References and provide a brief overview of the complement system and its ambiguous involvement in infections and other diseases.
-
-
- Lambris JD, editor. Current Topics in Complement. New York: Springer; 2006.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

