Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes
- PMID: 18188003
- PMCID: PMC2654259
- DOI: 10.4161/auto.5338
Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes
Abstract
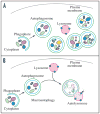
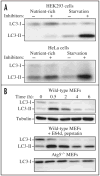
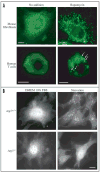
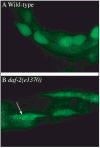
Research in autophagy continues to accelerate,(1) and as a result many new scientists are entering the field. Accordingly, it is important to establish a standard set of criteria for monitoring macroautophagy in different organisms. Recent reviews have described the range of assays that have been used for this purpose.(2,3) There are many useful and convenient methods that can be used to monitor macroautophagy in yeast, but relatively few in other model systems, and there is much confusion regarding acceptable methods to measure macroautophagy in higher eukaryotes. A key point that needs to be emphasized is that there is a difference between measurements that monitor the numbers of autophagosomes versus those that measure flux through the autophagy pathway; thus, a block in macroautophagy that results in autophagosome accumulation needs to be differentiated from fully functional autophagy that includes delivery to, and degradation within, lysosomes (in most higher eukaryotes) or the vacuole (in plants and fungi). Here, we present a set of guidelines for the selection and interpretation of the methods that can be used by investigators who are attempting to examine macroautophagy and related processes, as well as by reviewers who need to provide realistic and reasonable critiques of papers that investigate these processes. This set of guidelines is not meant to be a formulaic set of rules, because the appropriate assays depend in part on the question being asked and the system being used. In addition, we emphasize that no individual assay is guaranteed to be the most appropriate one in every situation, and we strongly recommend the use of multiple assays to verify an autophagic response.
Figures



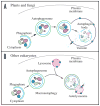

Comment in
-
Getting into the flow.Autophagy. 2008 Feb;4(2):139-40. doi: 10.4161/auto.5475. Epub 2007 Dec 28. Autophagy. 2008. PMID: 18188002 No abstract available.
Similar articles
-
Guidelines for the use and interpretation of assays for monitoring autophagy.Autophagy. 2012 Apr;8(4):445-544. doi: 10.4161/auto.19496. Autophagy. 2012. PMID: 22966490 Free PMC article.
-
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1.Autophagy. 2021 Jan;17(1):1-382. doi: 10.1080/15548627.2020.1797280. Epub 2021 Feb 8. Autophagy. 2021. PMID: 33634751 Free PMC article.
-
The correct way to monitor autophagy in higher eukaryotes.Autophagy. 2005 Jul;1(2):65. doi: 10.4161/auto.1.2.1899. Epub 2005 Jul 6. Autophagy. 2005. PMID: 16874029 No abstract available.
-
[Methods for monitoring macroautophagy].Tanpakushitsu Kakusan Koso. 2006 Aug;51(10 Suppl):1542-8. Tanpakushitsu Kakusan Koso. 2006. PMID: 16922435 Review. Japanese. No abstract available.
-
Autophagy: assays and artifacts.J Pathol. 2010 Jun;221(2):117-24. doi: 10.1002/path.2694. J Pathol. 2010. PMID: 20225337 Free PMC article. Review.
Cited by
-
Role of oxidative stress in cytotoxicity of grape seed extract in human bladder cancer cells.Food Chem Toxicol. 2013 Nov;61:187-95. doi: 10.1016/j.fct.2013.06.039. Epub 2013 Jul 3. Food Chem Toxicol. 2013. PMID: 23831192 Free PMC article.
-
EGFR tyrosine kinase inhibition induces autophagy in cancer cells.Cancer Biol Ther. 2012 Dec;13(14):1417-24. doi: 10.4161/cbt.22002. Epub 2012 Sep 6. Cancer Biol Ther. 2012. PMID: 22954701 Free PMC article.
-
Inhibition of autophagy impairs tumor cell invasion in an organotypic model.Cell Cycle. 2012 May 15;11(10):2022-9. doi: 10.4161/cc.20424. Epub 2012 May 15. Cell Cycle. 2012. PMID: 22580450 Free PMC article.
-
Use of ratiometrically designed nanocarrier targeting CDK4/6 and autophagy pathways for effective pancreatic cancer treatment.Nat Commun. 2020 Aug 25;11(1):4249. doi: 10.1038/s41467-020-17996-7. Nat Commun. 2020. PMID: 32843618 Free PMC article.
-
Effect of pantoprazole to enhance activity of docetaxel against human tumour xenografts by inhibiting autophagy.Br J Cancer. 2015 Mar 3;112(5):832-40. doi: 10.1038/bjc.2015.17. Epub 2015 Feb 3. Br J Cancer. 2015. Retraction in: Br J Cancer. 2024 Apr;130(7):1232. doi: 10.1038/s41416-024-02660-4 PMID: 25647012 Free PMC article. Retracted.
References
-
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. - PubMed
-
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. - PubMed
-
- Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–502. - PubMed
-
- Kovács AL, Reith A, Seglen PO. Accumulation of autophagosomes after inhibition of hepatocytic protein degradation by vinblastine, leupeptin or a lysosomotropic amine. Exp Cell Res. 1982;137:191–201. - PubMed
-
- Seglen PO. Regulation of autophagic protein degradation in isolated liver cells. In: Glaumann H, Ballard FJ, editors. Lysosomes: Their Role in Protein Breakdown. London: Academic Press; 1987. pp. 369–414.
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR055255-02/AR/NIAMS NIH HHS/United States
- TCR04004/TI_/Telethon/Italy
- R01 GM075061-03/GM/NIGMS NIH HHS/United States
- R01 GM053396/GM/NIGMS NIH HHS/United States
- R01 CA023378-29/CA/NCI NIH HHS/United States
- 064354/WT_/Wellcome Trust/United Kingdom
- R01 CA023378/CA/NCI NIH HHS/United States
- R01 GM075061/GM/NIGMS NIH HHS/United States
- R01 AI073099/AI/NIAID NIH HHS/United States
- G0600194/MRC_/Medical Research Council/United Kingdom
- R01 AG026389/AG/NIA NIH HHS/United States
- BB/E01030X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 AG026389-01A2/AG/NIA NIH HHS/United States
- R01 GM062509/GM/NIGMS NIH HHS/United States
- R01 AR055255/AR/NIAMS NIH HHS/United States
- G0601133/MRC_/Medical Research Council/United Kingdom
- G0600194(77639)/MRC_/Medical Research Council/United Kingdom
- GM53396/GM/NIGMS NIH HHS/United States
- R01 AI079065/AI/NIAID NIH HHS/United States
- R01 EY013520/EY/NEI NIH HHS/United States
- P01 AI056097/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
