Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells
- PMID: 18166975
- PMCID: PMC2692221
- DOI: 10.1038/cr.2008.8
Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells
Abstract
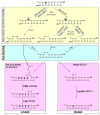
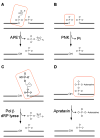
Base excision repair (BER) is an evolutionarily conserved process for maintaining genomic integrity by eliminating several dozen damaged (oxidized or alkylated) or inappropriate bases that are generated endogenously or induced by genotoxicants, predominantly, reactive oxygen species (ROS). BER involves 4-5 steps starting with base excision by a DNA glycosylase, followed by a common pathway usually involving an AP-endonuclease (APE) to generate 3' OH terminus at the damage site, followed by repair synthesis with a DNA polymerase and nick sealing by a DNA ligase. This pathway is also responsible for repairing DNA single-strand breaks with blocked termini directly generated by ROS. Nearly all glycosylases, far fewer than their substrate lesions particularly for oxidized bases, have broad and overlapping substrate range, and could serve as back-up enzymes in vivo. In contrast, mammalian cells encode only one APE, APE1, unlike two APEs in lower organisms. In spite of overall similarity, BER with distinct subpathways in the mammals is more complex than in E. coli. The glycosylases form complexes with downstream proteins to carry out efficient repair via distinct subpathways one of which, responsible for repair of strand breaks with 3' phosphate termini generated by the NEIL family glycosylases or by ROS, requires the phosphatase activity of polynucleotide kinase instead of APE1. Different complexes may utilize distinct DNA polymerases and ligases. Mammalian glycosylases have nonconserved extensions at one of the termini, dispensable for enzymatic activity but needed for interaction with other BER and non-BER proteins for complex formation and organelle targeting. The mammalian enzymes are sometimes covalently modified which may affect activity and complex formation. The focus of this review is on the early steps in mammalian BER for oxidized damage.
Figures
Similar articles
-
NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells.DNA Repair (Amst). 2006 Dec 9;5(12):1439-48. doi: 10.1016/j.dnarep.2006.07.003. Epub 2006 Sep 18. DNA Repair (Amst). 2006. PMID: 16982218 Free PMC article.
-
AP endonuclease-independent DNA base excision repair in human cells.Mol Cell. 2004 Jul 23;15(2):209-20. doi: 10.1016/j.molcel.2004.06.003. Mol Cell. 2004. PMID: 15260972
-
Oxidative DNA damage repair in mammalian cells: a new perspective.DNA Repair (Amst). 2007 Apr 1;6(4):470-80. doi: 10.1016/j.dnarep.2006.10.011. Epub 2006 Nov 20. DNA Repair (Amst). 2007. PMID: 17116430 Free PMC article. Review.
-
Study of interaction of XRCC1 with DNA and proteins of base excision repair by photoaffinity labeling technique.Biochemistry (Mosc). 2007 Aug;72(8):878-86. doi: 10.1134/s000629790708010x. Biochemistry (Mosc). 2007. PMID: 17922646
-
Oxidative genome damage and its repair: implications in aging and neurodegenerative diseases.Mech Ageing Dev. 2012 Apr;133(4):157-68. doi: 10.1016/j.mad.2012.01.005. Epub 2012 Jan 31. Mech Ageing Dev. 2012. PMID: 22313689 Free PMC article. Review.
Cited by
-
PARP-1 expression is increased in colon adenoma and carcinoma and correlates with OGG1.PLoS One. 2014 Dec 19;9(12):e115558. doi: 10.1371/journal.pone.0115558. eCollection 2014. PLoS One. 2014. PMID: 25526641 Free PMC article.
-
A Review of Numerical Models of Radiation Injury and Repair Considering Subcellular Targets and the Extracellular Microenvironment.Int J Mol Sci. 2024 Jan 13;25(2):1015. doi: 10.3390/ijms25021015. Int J Mol Sci. 2024. PMID: 38256089 Free PMC article. Review.
-
Replication Protein A Utilizes Differential Engagement of Its DNA-Binding Domains to Bind Biologically Relevant ssDNAs in Diverse Binding Modes.Biochemistry. 2022 Nov 15;61(22):2592-2606. doi: 10.1021/acs.biochem.2c00504. Epub 2022 Oct 24. Biochemistry. 2022. PMID: 36278947 Free PMC article.
-
The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes.J Biol Chem. 2013 May 10;288(19):13863-75. doi: 10.1074/jbc.M112.441444. Epub 2013 Mar 29. J Biol Chem. 2013. PMID: 23543741 Free PMC article.
-
The Mitochondrial Response to DNA Damage.Front Cell Dev Biol. 2021 May 12;9:669379. doi: 10.3389/fcell.2021.669379. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34055802 Free PMC article. Review.
References
-
- Dawson TL, Gores GJ, Nieminen AL, Herman B, Lemasters JJ. Mitochondria as a source of reactive oxygen species during reductive stress in rat hepatocytes. Am J Physiol. 1993;264:C961–967. - PubMed
-
- Griendling KK, Sorescu D, Ushio-Fukai M. NAD(P)H oxidase: role in cardiovascular biology and disease. Circ Res. 2000;86:494–501. - PubMed
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. - PubMed
-
- Wallace DJ. A reason for the rarity of male lupus. Lupus. 1998;7:60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

