The structure of the C-terminal actin-binding domain of talin
- PMID: 18157087
- PMCID: PMC2168396
- DOI: 10.1038/sj.emboj.7601965
The structure of the C-terminal actin-binding domain of talin
Abstract
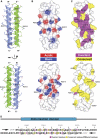
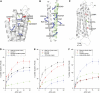
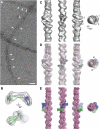
Talin is a large dimeric protein that couples integrins to cytoskeletal actin. Here, we report the structure of the C-terminal actin-binding domain of talin, the core of which is a five-helix bundle linked to a C-terminal helix responsible for dimerisation. The NMR structure of the bundle reveals a conserved surface-exposed hydrophobic patch surrounded by positively charged groups. We have mapped the actin-binding site to this surface and shown that helix 1 on the opposite side of the bundle negatively regulates actin binding. The crystal structure of the dimerisation helix reveals an antiparallel coiled-coil with conserved residues clustered on the solvent-exposed face. Mutagenesis shows that dimerisation is essential for filamentous actin (F-actin) binding and indicates that the dimerisation helix itself contributes to binding. We have used these structures together with small angle X-ray scattering to derive a model of the entire domain. Electron microscopy provides direct evidence for binding of the dimer to F-actin and indicates that it binds to three monomers along the long-pitch helix of the actin filament.
Figures


Similar articles
-
A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head.Structure. 2005 Jan;13(1):65-74. doi: 10.1016/j.str.2004.11.006. Structure. 2005. PMID: 15642262
-
Central region of talin has a unique fold that binds vinculin and actin.J Biol Chem. 2010 Sep 17;285(38):29577-87. doi: 10.1074/jbc.M109.095455. Epub 2010 Jul 7. J Biol Chem. 2010. PMID: 20610383 Free PMC article.
-
Characterization of an actin-binding site within the talin FERM domain.J Mol Biol. 2004 Oct 22;343(3):771-84. doi: 10.1016/j.jmb.2004.08.069. J Mol Biol. 2004. PMID: 15465061
-
The domain structure of talin: residues 1815-1973 form a five-helix bundle containing a cryptic vinculin-binding site.FEBS Lett. 2010 Jun 3;584(11):2237-41. doi: 10.1016/j.febslet.2010.04.028. Epub 2010 Apr 20. FEBS Lett. 2010. PMID: 20399778 Free PMC article.
-
Integrin connections to the cytoskeleton through talin and vinculin.Biochem Soc Trans. 2008 Apr;36(Pt 2):235-9. doi: 10.1042/BST0360235. Biochem Soc Trans. 2008. PMID: 18363566 Review.
Cited by
-
Vinculin activation is necessary for complete talin binding.Biophys J. 2011 Jan 19;100(2):332-40. doi: 10.1016/j.bpj.2010.11.024. Biophys J. 2011. PMID: 21244829 Free PMC article.
-
Talin 2 is a large and complex gene encoding multiple transcripts and protein isoforms.FEBS J. 2009 Mar;276(6):1610-28. doi: 10.1111/j.1742-4658.2009.06893.x. Epub 2009 Feb 7. FEBS J. 2009. PMID: 19220457 Free PMC article.
-
Structural Basis of β2 Integrin Inside-Out Activation.Cells. 2022 Sep 28;11(19):3039. doi: 10.3390/cells11193039. Cells. 2022. PMID: 36231001 Free PMC article. Review.
-
Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity.J Cell Biol. 2016 May 9;213(3):371-83. doi: 10.1083/jcb.201510012. J Cell Biol. 2016. PMID: 27161398 Free PMC article.
-
The structure of cell-matrix adhesions: the new frontier.Curr Opin Cell Biol. 2012 Feb;24(1):134-40. doi: 10.1016/j.ceb.2011.12.001. Epub 2011 Dec 22. Curr Opin Cell Biol. 2012. PMID: 22196929 Free PMC article. Review.
References
-
- Brett TJ, Legendre-Guillemin V, McPherson PS, Fremont DH (2006) Structural definition of the F-actin-binding THATCH domain from HIP1R. Nat Struct Mol Biol 13: 121–130 - PubMed
-
- Critchley DR (2004) Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem Soc Trans 32: 831–836 - PubMed
-
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 - PubMed
-
- Dominguez R (2004) Actin-binding proteins—a unifying hypothesis. Trends Biochem Sci 29: 572–578 - PubMed
-
- Egelman EH (2000) A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85: 225–234 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

