The cucumovirus 2b gene drives selection of inter-viral recombinants affecting the crossover site, the acceptor RNA and the rate of selection
- PMID: 18086712
- PMCID: PMC2275080
- DOI: 10.1093/nar/gkm1036
The cucumovirus 2b gene drives selection of inter-viral recombinants affecting the crossover site, the acceptor RNA and the rate of selection
Abstract
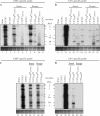
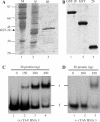
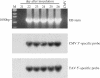
RNA-RNA recombination is an important pathway in virus evolution and has been described for many viruses. However, the factors driving recombination or promoting the selection of recombinants are still unclear. Here, we show that the small movement protein (2b) was able to promote selection of RNA 1/2-RNA 3 recombinants within a chimeric virus having RNAs 1 and 2 from cucumber mosaic virus, and RNA 3 from the related tomato aspermy virus, along with heterologous 2b genes. The source of the 2b also determined the selection of the acceptor RNA and the crossover site, as well as affecting the rate of selection of the recombinant RNAs. The nature of the RNA 3 also influenced the selection of the recombinant RNAs. A 163-nt tandem repeat in RNA 3 significantly affected the rate of selection of the recombinant RNA, while a single nucleotide within the repeat affected the crossover site. The recombination occurred in a non-random manner, involved no intermediates and probably was generated via a copy-choice mechanism during (+) strand RNA synthesis.
Figures




Similar articles
-
Recombination between genomic RNAs of two cucumoviruses under conditions of minimal selection pressure.Virology. 1999 Oct 25;263(2):282-9. doi: 10.1006/viro.1999.9973. Virology. 1999. PMID: 10544102
-
The 3' untranslated region of cucumber mosaic virus (CMV) subgroup II RNA3 arose by interspecific recombination between CMV and tomato aspermy virus.J Gen Virol. 2009 Sep;90(Pt 9):2293-8. doi: 10.1099/vir.0.011452-0. Epub 2009 May 27. J Gen Virol. 2009. PMID: 19474245
-
A map of the diversity of RNA3 recombinants appearing in plants infected with Cucumber mosaic virus and Tomato aspermy virus.Virology. 2005 Jan 5;331(1):117-27. doi: 10.1016/j.virol.2004.10.017. Virology. 2005. PMID: 15582658
-
Cucumoviruses.Adv Virus Res. 2003;62:241-323. doi: 10.1016/s0065-3527(03)62005-1. Adv Virus Res. 2003. PMID: 14719367 Review.
-
Molecular Modeling for Better Understanding of Cucumovirus Pathology.Adv Virus Res. 2018;102:59-88. doi: 10.1016/bs.aivir.2018.06.002. Epub 2018 Jul 26. Adv Virus Res. 2018. PMID: 30266176 Review.
Cited by
-
Generation and Retention of Defective RNA3 from Cucumber Mosaic Virus and Relevance of the 2b Protein to Generation of the Subviral RNA.Plant Pathol J. 2023 Dec;39(6):592-599. doi: 10.5423/PPJ.FT.07.2023.0106. Epub 2023 Dec 1. Plant Pathol J. 2023. PMID: 38081319 Free PMC article.
-
Fixation of emerging interviral recombinants in cucumber mosaic virus populations.J Virol. 2013 Jan;87(2):1264-9. doi: 10.1128/JVI.01892-12. Epub 2012 Oct 31. J Virol. 2013. PMID: 23115282 Free PMC article.
-
RNA binding is more critical to the suppression of silencing function of Cucumber mosaic virus 2b protein than nuclear localization.RNA. 2012 Apr;18(4):771-82. doi: 10.1261/rna.031260.111. Epub 2012 Feb 22. RNA. 2012. PMID: 22357910 Free PMC article.
-
Mapping viral functional domains for genetic diversity in plants.J Virol. 2013 Jan;87(2):790-7. doi: 10.1128/JVI.01891-12. Epub 2012 Oct 31. J Virol. 2013. PMID: 23115283 Free PMC article.
-
A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses.Biol Direct. 2012 Jun 11;7:13. doi: 10.1186/1745-6150-7-13. Biol Direct. 2012. PMID: 22515485 Free PMC article.
References
-
- Hirst GK. Genetic recombination with Newcastle disease virus, polioviruses, and influenza. Cold Spring Harb. Symp. Quant. Biol. 1962;27:303–309. - PubMed
-
- Ledinko N. Genetic recombination with poliovirus type 1. Studies of crosses between a normal horse serum-resistant mutant and several guanidine-resistant mutants of the same strain. Virology. 1963;20:107–119. - PubMed
-
- McCahon D. The genetics of aphthovirus. Brief review. Arch. Virol. 1981;69:1–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

