Analysis of the human polynucleotide phosphorylase (PNPase) reveals differences in RNA binding and response to phosphate compared to its bacterial and chloroplast counterparts
- PMID: 18083836
- PMCID: PMC2212259
- DOI: 10.1261/rna.698108
Analysis of the human polynucleotide phosphorylase (PNPase) reveals differences in RNA binding and response to phosphate compared to its bacterial and chloroplast counterparts
Abstract
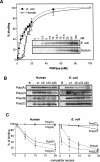
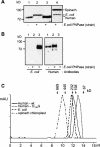
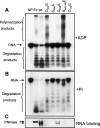
PNPase is a major exoribonuclease that plays an important role in the degradation, processing, and polyadenylation of RNA in prokaryotes and organelles. This phosphorolytic processive enzyme uses inorganic phosphate and nucleotide diphosphate for degradation and polymerization activities, respectively. Its structure and activities are similar to the archaeal exosome complex. The human PNPase was recently localized to the intermembrane space (IMS) of the mitochondria, and is, therefore, most likely not directly involved in RNA metabolism, unlike in bacteria and other organelles. In this work, the degradation, polymerization, and RNA-binding properties of the human PNPase were analyzed and compared to its bacterial and organellar counterparts. Phosphorolytic activity was displayed at lower optimum concentrations of inorganic phosphate. Also, the RNA-binding properties to ribohomopolymers varied significantly from those of its bacterial and organellar enzymes. The purified enzyme did not preferentially bind RNA harboring a poly(A) tail at the 3' end, compared to a molecule lacking this tail. Several site-directed mutations at conserved amino acid positions either eliminated or modified degradation/polymerization activity in different manners than observed for the Escherichia coli PNPase and the archaeal and human exosomes. In light of these results, a possible function of the human PNPase in the mitochondrial IMS is discussed.
Figures


Similar articles
-
Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins.Plant Cell. 2003 Sep;15(9):2003-19. doi: 10.1105/tpc.013326. Plant Cell. 2003. PMID: 12953107 Free PMC article.
-
Helicase SUV3, polynucleotide phosphorylase, and mitochondrial polyadenylation polymerase form a transient complex to modulate mitochondrial mRNA polyadenylated tail lengths in response to energetic changes.J Biol Chem. 2014 Jun 13;289(24):16727-35. doi: 10.1074/jbc.M113.536540. Epub 2014 Apr 25. J Biol Chem. 2014. PMID: 24770417 Free PMC article.
-
Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts.Mol Cell Biol. 2001 Aug;21(16):5408-16. doi: 10.1128/MCB.21.16.5408-5416.2001. Mol Cell Biol. 2001. PMID: 11463823 Free PMC article.
-
Running rings around RNA: a superfamily of phosphate-dependent RNases.Trends Biochem Sci. 2002 Jan;27(1):11-8. doi: 10.1016/s0968-0004(01)01999-5. Trends Biochem Sci. 2002. PMID: 11796219 Review.
-
Polynucleotide phosphorylase and the archaeal exosome as poly(A)-polymerases.Biochim Biophys Acta. 2008 Apr;1779(4):247-55. doi: 10.1016/j.bbagrm.2007.12.004. Epub 2007 Dec 15. Biochim Biophys Acta. 2008. PMID: 18177749 Review.
Cited by
-
Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA.RNA. 2008 Feb;14(2):310-23. doi: 10.1261/rna.697308. Epub 2007 Dec 14. RNA. 2008. PMID: 18083837 Free PMC article.
-
Inhibition of homologous phosphorolytic ribonucleases by citrate may represent an evolutionarily conserved communicative link between RNA degradation and central metabolism.Nucleic Acids Res. 2017 May 5;45(8):4655-4666. doi: 10.1093/nar/gkx114. Nucleic Acids Res. 2017. PMID: 28334892 Free PMC article.
-
Nonspherical Coacervate Shapes in an Enzyme-Driven Active System.Langmuir. 2020 Mar 3;36(8):1956-1964. doi: 10.1021/acs.langmuir.9b02719. Epub 2020 Feb 17. Langmuir. 2020. PMID: 31995710 Free PMC article.
-
Mitochondrial sequencing identifies long noncoding RNA features that promote binding to PNPase.Am J Physiol Cell Physiol. 2024 Aug 1;327(2):C221-C236. doi: 10.1152/ajpcell.00648.2023. Epub 2024 Jun 3. Am J Physiol Cell Physiol. 2024. PMID: 38826135
-
Mammalian mitochondrial RNAs are degraded in the mitochondrial intermembrane space by RNASET2.Protein Cell. 2017 Oct;8(10):735-749. doi: 10.1007/s13238-017-0448-9. Epub 2017 Jul 20. Protein Cell. 2017. PMID: 28730546 Free PMC article.
References
-
- Baginsky, S., Shteiman-Kotler, A., Liveanu, V., Yehudai-Resheff, S., Bellaoui, M., Settlage, R.E., Shabanowitz, J., Hunt, D.F., Schuster, G., Gruissem, W. Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA. 2001;7:1464–1475. - PMC - PubMed
-
- Bermudez-Cruz, R.M., Garcia-Mena, J., Montanez, C. Polynucleotide phosphorylase binds to ssRNA with same affinity as to ssDNA. Biochimie. 2002;84:321–328. - PubMed
-
- Buttner, K., Wenig, K., Hopfner, K.P. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell. 2005;20:461–471. - PubMed
-
- Buttner, K., Wenig, K., Hopfner, K.P. The exosome: A macromolecular cage for controlled RNA degradation. Mol. Microbiol. 2006;61:1372–1379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
