Biphasic incorporation of centromeric histone CENP-A in fission yeast
- PMID: 18077559
- PMCID: PMC2230595
- DOI: 10.1091/mbc.e07-05-0504
Biphasic incorporation of centromeric histone CENP-A in fission yeast
Abstract
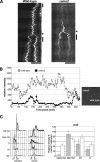
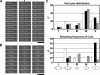
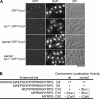
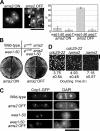
CENP-A is a centromere-specific histone H3 variant that is essential for kinetochore formation. Here, we report that the fission yeast Schizosaccharomyces pombe has at least two distinct CENP-A deposition phases across the cell cycle: S and G2. The S phase deposition requires Ams2 GATA factor, which promotes histone gene activation. In Delta ams2, CENP-A fails to retain during S, but it reaccumulates onto centromeres via the G2 deposition pathway, which is down-regulated by Hip1, a homologue of HIRA histone chaperon. Reducing the length of G2 in Delta ams2 results in failure of CENP-A accumulation, leading to chromosome missegregation. N-terminal green fluorescent protein-tagging reduces the centromeric association of CENP-A, causing cell death in Delta ams2 but not in wild-type cells, suggesting that the N-terminal tail of CENP-A may play a pivotal role in the formation of centromeric nucleosomes at G2. These observations imply that CENP-A is normally localized to centromeres in S phase in an Ams2-dependent manner and that the G2 pathway may salvage CENP-A assembly to promote genome stability. The flexibility of CENP-A incorporation during the cell cycle may account for the plasticity of kinetochore formation when the authentic centromere is damaged.
Figures

Similar articles
-
Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin.Mol Cell. 2009 Feb 13;33(3):287-98. doi: 10.1016/j.molcel.2009.01.017. Mol Cell. 2009. PMID: 19217403 Free PMC article.
-
Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast.J Biol Chem. 2010 Jan 15;285(3):1909-18. doi: 10.1074/jbc.M109.058487. Epub 2009 Nov 12. J Biol Chem. 2010. PMID: 19910462 Free PMC article.
-
Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP.Cell. 2009 May 1;137(3):472-84. doi: 10.1016/j.cell.2009.02.039. Cell. 2009. PMID: 19410544 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Posttranslational modifications of CENP-A: marks of distinction.Chromosoma. 2018 Sep;127(3):279-290. doi: 10.1007/s00412-018-0665-x. Epub 2018 Mar 22. Chromosoma. 2018. PMID: 29569072 Free PMC article. Review.
Cited by
-
Replicating centromeric chromatin: spatial and temporal control of CENP-A assembly.Exp Cell Res. 2012 Jul 15;318(12):1353-60. doi: 10.1016/j.yexcr.2012.04.007. Epub 2012 Apr 27. Exp Cell Res. 2012. PMID: 22561213 Free PMC article. Review.
-
Diversity in requirement of genetic and epigenetic factors for centromere function in fungi.Eukaryot Cell. 2011 Nov;10(11):1384-95. doi: 10.1128/EC.05165-11. Epub 2011 Sep 9. Eukaryot Cell. 2011. PMID: 21908596 Free PMC article. Review.
-
Rad51-Rad52 mediated maintenance of centromeric chromatin in Candida albicans.PLoS Genet. 2014 Apr 24;10(4):e1004344. doi: 10.1371/journal.pgen.1004344. eCollection 2014 Apr. PLoS Genet. 2014. PMID: 24762765 Free PMC article.
-
Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells.Cell. 2012 Oct 26;151(3):671-83. doi: 10.1016/j.cell.2012.09.019. Cell. 2012. PMID: 23101633 Free PMC article.
-
Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin.Mol Cell. 2009 Feb 13;33(3):299-311. doi: 10.1016/j.molcel.2009.01.019. Mol Cell. 2009. PMID: 19217404 Free PMC article.
References
-
- Ahmad K., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. - PubMed
-
- Black B. E., Foltz D. R., Chakravarthy S., Luger K., Woods V. L., Jr, Cleveland D. W. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. - PubMed
-
- Black B. E., Jansen L. E., Maddox P. S., Foltz D. R., Desai A. B., Shah J. V., Cleveland D. W. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007;25:309–322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

