Development of a mouse test for repetitive, restricted behaviors: relevance to autism
- PMID: 18068825
- PMCID: PMC2349090
- DOI: 10.1016/j.bbr.2007.10.029
Development of a mouse test for repetitive, restricted behaviors: relevance to autism
Abstract
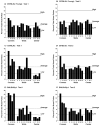
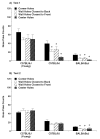
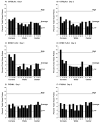
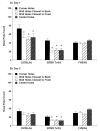
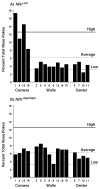
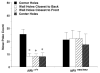
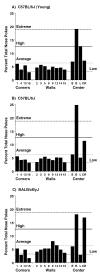
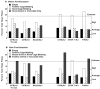
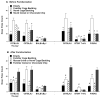
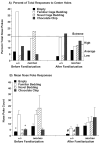
Repetitive behavior, a core symptom of autism, encompasses stereotyped responses, restricted interests, and resistance to change. These studies investigated whether different components of the repetitive behavior domain could be modeled in the exploratory hole-board task in mice. Four inbred mouse strains, C57BL/6J, BALB/cByJ, BTBR T+tf/J, and FVB/NJ, and mice with reduced expression of Grin1, leading to NMDA receptor hypofunction (NR1neo/neo mice), were tested for exploration and preference for olfactory stimuli in an activity chamber with a 16-hole floor-board. Reduced exploration and high preference for holes located in the corners of the chamber were observed in BALB/cByJ and BTBR T+tf/J mice. All inbred strains had initial high preference for a familiar olfactory stimulus (clean cage bedding). BTBR T+tf/J was the only strain that did not demonstrate a shift in hole preference towards an appetitive olfactory stimulus (cereal or a chocolate chip), following home cage exposure to the food. The NR1neo/neo mice showed lower hole selectivity and aberrant olfactory stimulus preference, in comparison to wildtype controls. The results indicate that NR1neo/neo mice have repetitive nose poke responses that are less modified by environmental contingencies than responses in wildtype mice. 25-30% of NMDA receptor hypomorphic mice also show self-injurious responses. Findings from the olfactory studies suggest that resistance to change and restricted interests might be modeled in mice by a failure to alter patterns of hole preference following familiarization with an appetitive stimulus, and by high preference persistently demonstrated for one particular olfactory stimulus. Further work is required to determine the characteristics of optimal mouse social stimuli in the olfactory hole-board test.
Figures

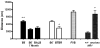
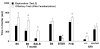
Similar articles
-
Repetitive behavior profile and supersensitivity to amphetamine in the C58/J mouse model of autism.Behav Brain Res. 2014 Feb 1;259:200-14. doi: 10.1016/j.bbr.2013.10.052. Epub 2013 Nov 8. Behav Brain Res. 2014. PMID: 24211371 Free PMC article.
-
Inbred strain preference in the BTBR T+ Itpr3tf /J mouse model of autism spectrum disorder: Does the stranger mouse matter in social approach?Autism Res. 2019 Aug;12(8):1184-1191. doi: 10.1002/aur.2158. Epub 2019 Jun 17. Autism Res. 2019. PMID: 31206258
-
Mouse behavioral tasks relevant to autism: phenotypes of 10 inbred strains.Behav Brain Res. 2007 Jan 10;176(1):4-20. doi: 10.1016/j.bbr.2006.07.030. Epub 2006 Sep 12. Behav Brain Res. 2007. PMID: 16971002 Free PMC article.
-
Mouse behavioral assays relevant to the symptoms of autism.Brain Pathol. 2007 Oct;17(4):448-59. doi: 10.1111/j.1750-3639.2007.00096.x. Brain Pathol. 2007. PMID: 17919130 Free PMC article. Review.
-
The Hole-Board Test in Mutant Mice.Behav Genet. 2022 May;52(3):158-169. doi: 10.1007/s10519-022-10102-1. Epub 2022 Apr 28. Behav Genet. 2022. PMID: 35482162 Review.
Cited by
-
Hippocampal neuroligin-2 overexpression leads to reduced aggression and inhibited novelty reactivity in rats.PLoS One. 2013;8(2):e56871. doi: 10.1371/journal.pone.0056871. Epub 2013 Feb 22. PLoS One. 2013. PMID: 23451101 Free PMC article.
-
Preweaning sensorimotor deficits and adolescent hypersociability in Grin1 knockdown mice.Dev Neurosci. 2012;34(2-3):159-73. doi: 10.1159/000337984. Epub 2012 May 8. Dev Neurosci. 2012. PMID: 22571986 Free PMC article.
-
Molecular Abnormalities in BTBR Mice and Their Relevance to Schizophrenia and Autism Spectrum Disorders: An Overview of Transcriptomic and Proteomic Studies.Biomedicines. 2023 Jan 20;11(2):289. doi: 10.3390/biomedicines11020289. Biomedicines. 2023. PMID: 36830826 Free PMC article.
-
The Absence of Caspase-8 in the Dopaminergic System Leads to Mild Autism-like Behavior.Front Cell Dev Biol. 2022 Apr 5;10:839715. doi: 10.3389/fcell.2022.839715. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35493109 Free PMC article.
-
Modeling rare gene variation to gain insight into the oldest biomarker in autism: construction of the serotonin transporter Gly56Ala knock-in mouse.J Neurodev Disord. 2009 Jun;1(2):158-71. doi: 10.1007/s11689-009-9020-0. J Neurodev Disord. 2009. PMID: 19960097 Free PMC article.
References
-
- Alexander AL, Lee JE, Lazar M, Boudos R, Dubray MB, Oakes TR, Miller JN, Lu J, Jeong EK, McMahon WM, Bigler ED, Lainhart JE. Diffusion tensor imaging of the corpus callosum in Autism. Neuroimage. 2007;34:61–73. - PubMed
-
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) American Psychiatric Publishing, Inc; Washington, DC: 2000.
-
- Amico JA, Vollmer RR, Cai HM, Miedlar JA, Rinaman L. Enhanced initial and sustained intake of sucrose solution in mice with an oxytocin gene deletion. Am J Physiol Regul Integr Comp Physiol. 2005;289:R1798–1806. - PubMed
-
- Asaka Y, Jugloff DG, Zhang L, Eubanks JH, Fitzsimonds RM. Hippocampal synaptic plasticity is impaired in the Mecp 2-null mouse model of Rett syndrome. Neurobiol Dis. 2006;21:217–227. - PubMed
-
- Aylward EH, Minshew NJ, Goldstein G, Honeycutt NA, Augustine AM, Yates KO, Barta PE, Pearlson GD. MRI volumes of amygdala and hippocampus in non-mentally retarded autistic adolescents and adults. Neurology. 1999;53:2145–2150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 HD003110-40/HD/NICHD NIH HHS/United States
- MH063398/MH/NIMH NIH HHS/United States
- U54 MH066418/MH/NIMH NIH HHS/United States
- R01 MH063398-05/MH/NIMH NIH HHS/United States
- U54 MH066418-05/MH/NIMH NIH HHS/United States
- R01 MH73402/MH/NIMH NIH HHS/United States
- U54 MH66418/MH/NIMH NIH HHS/United States
- P30 HD03110/HD/NICHD NIH HHS/United States
- P30 HD003110/HD/NICHD NIH HHS/United States
- R01 MH073402-04/MH/NIMH NIH HHS/United States
- R01 MH063398/MH/NIMH NIH HHS/United States
- R01 MH073402/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

