Genome-wide view of cell fate specification: ladybird acts at multiple levels during diversification of muscle and heart precursors
- PMID: 18056427
- PMCID: PMC2081981
- DOI: 10.1101/gad.437307
Genome-wide view of cell fate specification: ladybird acts at multiple levels during diversification of muscle and heart precursors
Abstract
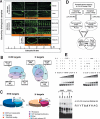
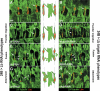
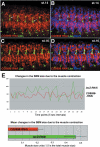
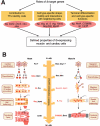
Correct diversification of cell types during development ensures the formation of functional organs. The evolutionarily conserved homeobox genes from ladybird/Lbx family were found to act as cell identity genes in a number of embryonic tissues. A prior genetic analysis showed that during Drosophila muscle and heart development ladybird is required for the specification of a subset of muscular and cardiac precursors. To learn how ladybird genes exert their cell identity functions we performed muscle and heart-targeted genome-wide transcriptional profiling and a chromatin immunoprecipitation (ChIP)-on-chip search for direct Ladybird targets. Our data reveal that ladybird not only contributes to the combinatorial code of transcription factors specifying the identity of muscle and cardiac precursors, but also regulates a large number of genes involved in setting cell shape, adhesion, and motility. Among direct ladybird targets, we identified bric-a-brac 2 gene as a new component of identity code and inflated encoding alphaPS2-integrin playing a pivotal role in cell-cell interactions. Unexpectedly, ladybird also contributes to the regulation of terminal differentiation genes encoding structural muscle proteins or contributing to muscle contractility. Thus, the identity gene-governed diversification of cell types is a multistep process involving the transcriptional control of genes determining both morphological and functional properties of cells.
Figures


Similar articles
-
Modifiers of muscle and heart cell fate specification identified by gain-of-function screen in Drosophila.Mech Dev. 2003 Sep;120(9):991-1007. doi: 10.1016/s0925-4773(03)00182-5. Mech Dev. 2003. PMID: 14550529
-
ladybird, a new component of the cardiogenic pathway in Drosophila required for diversification of heart precursors.Development. 1997 Sep;124(18):3471-9. doi: 10.1242/dev.124.18.3471. Development. 1997. PMID: 9342040
-
Shaping leg muscles in Drosophila: role of ladybird, a conserved regulator of appendicular myogenesis.PLoS One. 2006 Dec 27;1(1):e122. doi: 10.1371/journal.pone.0000122. PLoS One. 2006. PMID: 17205126 Free PMC article.
-
A cluster of Drosophila homeobox genes involved in mesoderm differentiation programs.Bioessays. 2001 Feb;23(2):125-33. doi: 10.1002/1521-1878(200102)23:2<125::AID-BIES1019>3.0.CO;2-C. Bioessays. 2001. PMID: 11169585 Review.
-
Acting on identity: Myoblast fusion and the formation of the syncytial muscle fiber.Semin Cell Dev Biol. 2017 Dec;72:45-55. doi: 10.1016/j.semcdb.2017.10.033. Epub 2017 Nov 6. Semin Cell Dev Biol. 2017. PMID: 29101004 Free PMC article. Review.
Cited by
-
Inhibiting stromal cell heparan sulfate synthesis improves stem cell mobilization and enables engraftment without cytotoxic conditioning.Blood. 2014 Nov 6;124(19):2937-47. doi: 10.1182/blood-2014-08-593426. Epub 2014 Sep 8. Blood. 2014. PMID: 25202142 Free PMC article.
-
Specification of the somatic musculature in Drosophila.Wiley Interdiscip Rev Dev Biol. 2015 Jul-Aug;4(4):357-75. doi: 10.1002/wdev.182. Epub 2015 Feb 27. Wiley Interdiscip Rev Dev Biol. 2015. PMID: 25728002 Free PMC article. Review.
-
Genetic networks governing heart development.Cold Spring Harb Perspect Med. 2014 Oct 3;4(11):a013839. doi: 10.1101/cshperspect.a013839. Cold Spring Harb Perspect Med. 2014. PMID: 25280899 Free PMC article. Review.
-
A cell atlas of adult muscle precursors uncovers early events in fibre-type divergence in Drosophila.EMBO Rep. 2020 Oct 5;21(10):e49555. doi: 10.15252/embr.201949555. Epub 2020 Aug 19. EMBO Rep. 2020. PMID: 32815271 Free PMC article.
-
Genetic control of muscle development: learning from Drosophila.J Muscle Res Cell Motil. 2007;28(7-8):397-407. doi: 10.1007/s10974-008-9133-1. Epub 2008 Mar 18. J Muscle Res Cell Motil. 2007. PMID: 18347920 Review.
References
-
- Alvares L.E., Schubert F.R., Thorpe C., Mootoosamy R.C., Cheng L., Parkyn G., Lumsden A., Dietrich S., Schubert F.R., Thorpe C., Mootoosamy R.C., Cheng L., Parkyn G., Lumsden A., Dietrich S., Thorpe C., Mootoosamy R.C., Cheng L., Parkyn G., Lumsden A., Dietrich S., Mootoosamy R.C., Cheng L., Parkyn G., Lumsden A., Dietrich S., Cheng L., Parkyn G., Lumsden A., Dietrich S., Parkyn G., Lumsden A., Dietrich S., Lumsden A., Dietrich S., Dietrich S. Intrinsic, Hox-dependent cues determine the fate of skeletal muscle precursors. Dev. Cell. 2003;3:379–390. - PubMed
-
- Baylies M.K., Michelson A.M., Michelson A.M. Invertebrate myogenesis: Looking back to the future of muscle development. Curr. Opin. Genet. Dev. 2001;11:431–439. - PubMed
-
- Briscoe J., Pierani A., Jessell T.M., Ericson J., Pierani A., Jessell T.M., Ericson J., Jessell T.M., Ericson J., Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. - PubMed
-
- Brohmann H., Jagla K., Birchmeier C., Jagla K., Birchmeier C., Birchmeier C. The role of Lbx1 in migration of muscle precursor cells. Development. 2000;127:437–445. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
