Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis
- PMID: 18056411
- PMCID: PMC2099190
- DOI: 10.1083/jcb.200706034
Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis
Abstract
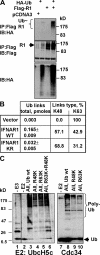
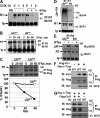
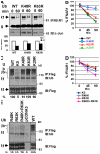
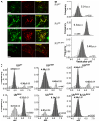
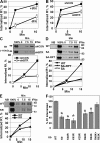
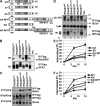
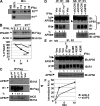
Ligand-induced endocytosis and lysosomal degradation of cognate receptors regulate the extent of cell signaling. Along with linear endocytic motifs that recruit the adaptin protein complex 2 (AP2)-clathrin molecules, monoubiquitination of receptors has emerged as a major endocytic signal. By investigating ubiquitin-dependent lysosomal degradation of the interferon (IFN)-alpha/beta receptor 1 (IFNAR1) subunit of the type I IFN receptor, we reveal that IFNAR1 is polyubiquitinated via both Lys48- and Lys63-linked chains. The SCF(betaTrcp) (Skp1-Cullin1-F-box complex) E3 ubiquitin ligase that mediates IFNAR1 ubiquitination and degradation in cells can conjugate both types of chains in vitro. Although either polyubiquitin linkage suffices for postinternalization sorting, both types of chains are necessary but not sufficient for robust IFNAR1 turnover and internalization. These processes also depend on the proximity of ubiquitin-acceptor lysines to a linear endocytic motif and on its integrity. Furthermore, ubiquitination of IFNAR1 promotes its interaction with the AP2 adaptin complex that is required for the robust internalization of IFNAR1, implicating cooperation between site-specific ubiquitination and the linear endocytic motif in regulating this process.
Figures
Similar articles
-
Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor.J Biol Chem. 2004 Nov 5;279(45):46614-20. doi: 10.1074/jbc.M407082200. Epub 2004 Aug 26. J Biol Chem. 2004. PMID: 15337770
-
SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor.EMBO J. 2003 Oct 15;22(20):5480-90. doi: 10.1093/emboj/cdg524. EMBO J. 2003. PMID: 14532120 Free PMC article.
-
Basal ubiquitin-independent internalization of interferon alpha receptor is prevented by Tyk2-mediated masking of a linear endocytic motif.J Biol Chem. 2008 Jul 4;283(27):18566-72. doi: 10.1074/jbc.M800991200. Epub 2008 May 12. J Biol Chem. 2008. PMID: 18474601 Free PMC article.
-
Interferon Receptor Trafficking and Signaling: Journey to the Cross Roads.Front Immunol. 2021 Jan 20;11:615603. doi: 10.3389/fimmu.2020.615603. eCollection 2020. Front Immunol. 2021. PMID: 33552080 Free PMC article. Review.
-
The role of K63-linked polyubiquitin in several types of autophagy.Biol Futur. 2022 Jun;73(2):137-148. doi: 10.1007/s42977-022-00117-4. Epub 2022 May 2. Biol Futur. 2022. PMID: 35501575 Review.
Cited by
-
Ubiquitination-mediated regulation of interferon responses.Growth Factors. 2012 Jun;30(3):141-8. doi: 10.3109/08977194.2012.669382. Epub 2012 Mar 7. Growth Factors. 2012. PMID: 22394219 Free PMC article. Review.
-
Roles and functions of IAV proteins in host immune evasion.Front Immunol. 2023 Dec 13;14:1323560. doi: 10.3389/fimmu.2023.1323560. eCollection 2023. Front Immunol. 2023. PMID: 38152399 Free PMC article. Review.
-
BRCA1-A and BRISC: Multifunctional Molecular Machines for Ubiquitin Signaling.Biomolecules. 2020 Oct 31;10(11):1503. doi: 10.3390/biom10111503. Biomolecules. 2020. PMID: 33142801 Free PMC article. Review.
-
Three ubiquitination sites of organic anion transporter-1 synergistically mediate protein kinase C-dependent endocytosis of the transporter.Mol Pharmacol. 2013 Jul;84(1):139-46. doi: 10.1124/mol.113.086769. Epub 2013 May 2. Mol Pharmacol. 2013. PMID: 23640180 Free PMC article.
-
Protein tyrosine phosphatase 1B is a key regulator of IFNAR1 endocytosis and a target for antiviral therapies.Proc Natl Acad Sci U S A. 2012 Nov 20;109(47):19226-31. doi: 10.1073/pnas.1211491109. Epub 2012 Nov 5. Proc Natl Acad Sci U S A. 2012. PMID: 23129613 Free PMC article.
References
-
- Barriere, H., C. Nemes, D. Lechardeur, M. Khan-Mohammad, K. Fruh, and G.L. Lukacs. 2006. Molecular basis of oligoubiquitin-dependent internalization of membrane proteins in mammalian cells. Traffic. 7:282–297. - PubMed
-
- Bonifacino, J.S., and L.M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447. - PubMed
-
- Brzovic, P.S., and R.E. Klevit. 2006. Ubiquitin transfer from the E2 perspective: why is UbcH5 so promiscuous? Cell Cycle. 5:2867–2873. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

