HIV-1 matrix protein p17 binds to monocytes and selectively stimulates MCP-1 secretion: role of transcriptional factor AP-1
- PMID: 18042260
- PMCID: PMC7162350
- DOI: 10.1111/j.1462-5822.2007.01073.x
HIV-1 matrix protein p17 binds to monocytes and selectively stimulates MCP-1 secretion: role of transcriptional factor AP-1
Abstract
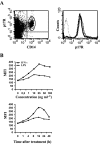
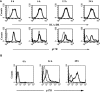
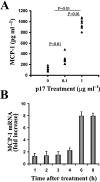
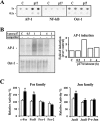
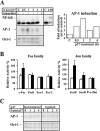
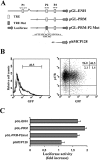
HIV-1 matrix protein p17 activates a variety of cell responses which play a critical role in viral replication and infection. Its activity depends on the expression of p17 receptors (p17R) on the surface of target cells. Whether p17 also plays a role in stimulating human monocytes, a major HIV-1 reservoir, is not known. Here we show that human monocytes constitutively express p17Rs and that p17 selectively triggers these cells to produce MCP-1. The effect of p17 on MCP-1 expression was observed at the transcriptional level and was primarily dependent on the activation of the transcription factor AP-1. p17 increased the binding activity of AP-1 complexes in a time- and dose-dependent manner. Deletion of the AP-1 binding sites in the MCP-1 promoter resulted in the lack of p17-induced MCP-1 transcription. In particular, the P3 binding site located between -69 and -63 position seems to be essential to MCP-1 mRNA induction in p17-treated monocytes. An ever increasing amount of evidences shows a tight link between biologically dysregulated monocytes, AP-1 activation, MCP-1 release and HIV-1 pathogenesis. Overall our results suggest that p17 may play a critical role in the monocyte-mediated inflammatory processes, which are suspected to be major precipitating events in AIDS-defining diseases.
Figures
Similar articles
-
Opposite effects of HIV-1 p17 variants on PTEN activation and cell growth in B cells.PLoS One. 2011 Mar 14;6(3):e17831. doi: 10.1371/journal.pone.0017831. PLoS One. 2011. PMID: 21423810 Free PMC article.
-
Simian immunodeficiency virus and human immunodeficiency virus type 1 matrix proteins specify different capabilities to modulate B cell growth.J Virol. 2014 May;88(10):5706-17. doi: 10.1128/JVI.03142-13. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623414 Free PMC article.
-
HIV-1 matrix protein p17 binds to the IL-8 receptor CXCR1 and shows IL-8-like chemokine activity on monocytes through Rho/ROCK activation.Blood. 2012 Mar 8;119(10):2274-83. doi: 10.1182/blood-2011-06-364083. Epub 2012 Jan 18. Blood. 2012. PMID: 22262769
-
HIV-1 Matrix Protein p17 and its Receptors.Curr Drug Targets. 2016;17(1):23-32. doi: 10.2174/1389450116666150825110840. Curr Drug Targets. 2016. PMID: 26302809 Review.
-
Functions of the HIV-1 matrix protein p17.New Microbiol. 2006 Jan;29(1):1-10. New Microbiol. 2006. PMID: 16608119 Review.
Cited by
-
The HIV-1 Matrix Protein p17 Does Cross the Blood-Brain Barrier.J Virol. 2022 Jan 12;96(1):e0120021. doi: 10.1128/JVI.01200-21. Epub 2021 Oct 20. J Virol. 2022. PMID: 34668776 Free PMC article.
-
Cellular aspartyl proteases promote the unconventional secretion of biologically active HIV-1 matrix protein p17.Sci Rep. 2016 Dec 1;6:38027. doi: 10.1038/srep38027. Sci Rep. 2016. PMID: 27905556 Free PMC article.
-
The cytokine/chemokine response in Leishmania/HIV infection and co-infection.Heliyon. 2023 Apr 1;9(4):e15055. doi: 10.1016/j.heliyon.2023.e15055. eCollection 2023 Apr. Heliyon. 2023. PMID: 37082641 Free PMC article. Review.
-
HIV interactions with monocytes and dendritic cells: viral latency and reservoirs.Retrovirology. 2009 Jun 1;6:51. doi: 10.1186/1742-4690-6-51. Retrovirology. 2009. PMID: 19486514 Free PMC article. Review.
-
HIV-1 matrix protein p17 promotes angiogenesis via chemokine receptors CXCR1 and CXCR2.Proc Natl Acad Sci U S A. 2012 Sep 4;109(36):14580-5. doi: 10.1073/pnas.1206605109. Epub 2012 Aug 17. Proc Natl Acad Sci U S A. 2012. PMID: 22904195 Free PMC article.
References
-
- Alfano, M. , and Poli, G. (2001) Cytokine and chemokine based control of HIV infection and replication. Curr Pharm Des 7: 993–1013. - PubMed
-
- Behren, A. , Simon, C. , Schwab, B.M. , Loetzsch, E. , Brodbeck, S. , Huber, E. , et al. (2005) Papillomavirus E2 protein induces expression of the matrix metalloproteinase‐9 via the extracellular signal‐regulated kinase/activator protein‐1 pathway. Cancer Res 65: 11613–11621. - PubMed
-
- Biggs, T.E. , Cooke, S.J. , Barton, C.H. , Harris, M.P. , Saksela, K. , and Mann, D.A. (1999) Induction of activator protein 1 (AP‐1) in macrophages by human immunodeficiency virus type‐1 NEF is a cell‐type‐specific response that requires both hck and MAPK signalling events. J Mol Biol 290: 21–35. - PubMed
-
- Cho, N.‐H. , Seong, S.‐Y. , Huh, M.‐S. , Kim, N.‐H. , Choi, M.‐S. , and Kim, I.‐S. (2002) Induction of the gene encoding macrophage chemoattractant protein 1 by Orientia tsutsugamushi in human endothelial cells involves activation of transcription factor activator protein 1. Infect Immun 70: 4841–4850. - PMC - PubMed
-
- Cinque, P. , Vago, L. , Mengozzi, M. , Torri, V. , Ceresa, D. , Licenzi, E. , et al. (1998) Elevated cerebrospinal fluid levels of monocyte chemotactic protein‐1 correlate with HIV‐1 encephalitis and local viral replication. AIDS 12: 1327–1332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

