Organism size promotes the evolution of specialized cells in multicellular digital organisms
- PMID: 18036158
- PMCID: PMC2387225
- DOI: 10.1111/j.1420-9101.2007.01466.x
Organism size promotes the evolution of specialized cells in multicellular digital organisms
Abstract
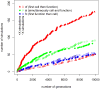
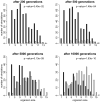
Specialized cells are the essence of complex multicellular life. Fossils allow us to study the modification of specialized, multicellular features such as jaws, scales, and muscular appendages. But it is still unclear what organismal properties contributed to the transition from undifferentiated organisms, which contain only a single cell type, to multicellular organisms with specialized cells. Using digital organisms I studied this transition. My simulations show that the transition to specialized cells happens faster in organism composed of many cells than in organisms composed of few cells. Large organisms suffer less from temporarily unsuccessful evolutionary experiments with individual cells, allowing them to evolve specialized cells via evolutionary trajectories that are unavailable to smaller organisms. This demonstrates that the evolution of simple multicellular organisms which are composed of many functionally identical cells accelerates the evolution of more complex organisms with specialized cells.
Figures


Similar articles
-
An evolutionary scenario for the transition to undifferentiated multicellularity.Proc Natl Acad Sci U S A. 2003 Feb 4;100(3):1095-8. doi: 10.1073/pnas.0335420100. Epub 2003 Jan 23. Proc Natl Acad Sci U S A. 2003. PMID: 12547910 Free PMC article.
-
Germ lines and extended selection during the evolutionary transition to multicellularity.J Exp Zool B Mol Dev Evol. 2021 Dec;336(8):680-686. doi: 10.1002/jez.b.22985. Epub 2020 Jul 18. J Exp Zool B Mol Dev Evol. 2021. PMID: 32681710
-
The evolutionary origin of somatic cells under the dirty work hypothesis.PLoS Biol. 2014 May 13;12(5):e1001858. doi: 10.1371/journal.pbio.1001858. eCollection 2014 May. PLoS Biol. 2014. PMID: 24823361 Free PMC article.
-
Origin of multicellular organisms as an inevitable consequence of dynamical systems.Anat Rec. 2002 Nov 1;268(3):327-42. doi: 10.1002/ar.10164. Anat Rec. 2002. PMID: 12382328 Review.
-
Mosaic physiology from developmental noise: within-organism physiological diversity as an alternative to phenotypic plasticity and phenotypic flexibility.J Exp Biol. 2014 Jan 1;217(Pt 1):35-45. doi: 10.1242/jeb.089698. J Exp Biol. 2014. PMID: 24353202 Review.
Cited by
-
A process engineering approach to increase organoid yield.Development. 2017 Mar 15;144(6):1128-1136. doi: 10.1242/dev.142919. Epub 2017 Feb 7. Development. 2017. PMID: 28174251 Free PMC article.
-
Experimental evolution of multicellularity.Proc Natl Acad Sci U S A. 2012 Jan 31;109(5):1595-600. doi: 10.1073/pnas.1115323109. Epub 2012 Jan 17. Proc Natl Acad Sci U S A. 2012. PMID: 22307617 Free PMC article.
-
Evolution of reproductive development in the volvocine algae.Sex Plant Reprod. 2011 Jun;24(2):97-112. doi: 10.1007/s00497-010-0158-4. Epub 2010 Dec 21. Sex Plant Reprod. 2011. PMID: 21174128 Free PMC article. Review.
-
From Species to Regional and Local Specialization of Intestinal Macrophages.Front Cell Dev Biol. 2021 Feb 18;8:624213. doi: 10.3389/fcell.2020.624213. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33681185 Free PMC article. Review.
-
Topological constraints in early multicellularity favor reproductive division of labor.Elife. 2020 Sep 17;9:e54348. doi: 10.7554/eLife.54348. Elife. 2020. PMID: 32940598 Free PMC article.
References
-
- Bell G, Mooers A. Size and complexity among multicellular organisms. Biol J Linnean Soc. 1997;60:345–363.
-
- Blau P. On the nature of organizations. Wiley; New York: 1974.
-
- Bonner JT. Perspective: the size-complexity rule. Evolution Int J Org Evolution. 2004;58:1883–90. - PubMed
-
- Bonner JT. Size and Cycle. Princeton University Press; 1965.

