Dendroscope: An interactive viewer for large phylogenetic trees
- PMID: 18034891
- PMCID: PMC2216043
- DOI: 10.1186/1471-2105-8-460
Dendroscope: An interactive viewer for large phylogenetic trees
Abstract
Background: Research in evolution requires software for visualizing and editing phylogenetic trees, for increasingly very large datasets, such as arise in expression analysis or metagenomics, for example. It would be desirable to have a program that provides these services in an efficient and user-friendly way, and that can be easily installed and run on all major operating systems. Although a large number of tree visualization tools are freely available, some as a part of more comprehensive analysis packages, all have drawbacks in one or more domains. They either lack some of the standard tree visualization techniques or basic graphics and editing features, or they are restricted to small trees containing only tens of thousands of taxa. Moreover, many programs are difficult to install or are not available for all common operating systems.
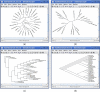
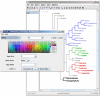
Results: We have developed a new program, Dendroscope, for the interactive visualization and navigation of phylogenetic trees. The program provides all standard tree visualizations and is optimized to run interactively on trees containing hundreds of thousands of taxa. The program provides tree editing and graphics export capabilities. To support the inspection of large trees, Dendroscope offers a magnification tool. The software is written in Java 1.4 and installers are provided for Linux/Unix, MacOS X and Windows XP.
Conclusion: Dendroscope is a user-friendly program for visualizing and navigating phylogenetic trees, for both small and large datasets.
Figures

Similar articles
-
TreeQ-VISTA: an interactive tree visualization tool with functional annotation query capabilities.Bioinformatics. 2007 Mar 15;23(6):764-6. doi: 10.1093/bioinformatics/btl643. Epub 2007 Jan 17. Bioinformatics. 2007. PMID: 17234642
-
Visualizing phylogenetic trees using TreeView.Curr Protoc Bioinformatics. 2002 Aug;Chapter 6:Unit 6.2. doi: 10.1002/0471250953.bi0602s01. Curr Protoc Bioinformatics. 2002. PMID: 18792942
-
Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks.Syst Biol. 2012 Dec 1;61(6):1061-7. doi: 10.1093/sysbio/sys062. Epub 2012 Jul 10. Syst Biol. 2012. PMID: 22780991
-
PET/CT image navigation and communication.J Nucl Med. 2004 Jan;45 Suppl 1:46S-55S. J Nucl Med. 2004. PMID: 14736835 Review.
-
A road map for phylogenetic models of species trees.Mol Phylogenet Evol. 2022 Aug;173:107483. doi: 10.1016/j.ympev.2022.107483. Epub 2022 Apr 30. Mol Phylogenet Evol. 2022. PMID: 35500743 Review.
Cited by
-
General patterns of diversity in major marine microeukaryote lineages.PLoS One. 2013;8(2):e57170. doi: 10.1371/journal.pone.0057170. Epub 2013 Feb 21. PLoS One. 2013. PMID: 23437337 Free PMC article.
-
Structure-based design, synthesis, and characterization of dual hotspot small-molecule HIV-1 entry inhibitors.J Med Chem. 2012 May 10;55(9):4382-96. doi: 10.1021/jm300265j. Epub 2012 Apr 23. J Med Chem. 2012. PMID: 22497421 Free PMC article.
-
Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine.Front Plant Sci. 2012 Feb 7;3:19. doi: 10.3389/fpls.2012.00019. eCollection 2012. Front Plant Sci. 2012. PMID: 22639640 Free PMC article.
-
Comparative analysis of RNA families reveals distinct repertoires for each domain of life.PLoS Comput Biol. 2012;8(11):e1002752. doi: 10.1371/journal.pcbi.1002752. Epub 2012 Nov 1. PLoS Comput Biol. 2012. PMID: 23133357 Free PMC article.
-
Resolution and characterization of distinct cpn60-based subgroups of Gardnerella vaginalis in the vaginal microbiota.PLoS One. 2012;7(8):e43009. doi: 10.1371/journal.pone.0043009. Epub 2012 Aug 10. PLoS One. 2012. PMID: 22900080 Free PMC article.
References
-
- Maddison DR, Schulz KS. The Tree of Life Web Project. 2006. http://tolweb.org
-
- Munzner T, Guimbretière F, Tasiran S, Zhang L, Zhou Y. TreeJuxtaposer: scalable tree comparison using Focus+Context with guaranteed visibility. ACM Trans Graph. 2003;22:453–462. doi: 10.1145/882262.882291. - DOI
-
- Felsenstein J. Tree plotting/drawing software. 2007. http://evolution.genetics.washington.edu/phylip/software.html#Plotting
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

