Copper binding to the Alzheimer's disease amyloid precursor protein
- PMID: 18030462
- PMCID: PMC2921068
- DOI: 10.1007/s00249-007-0234-3
Copper binding to the Alzheimer's disease amyloid precursor protein
Abstract
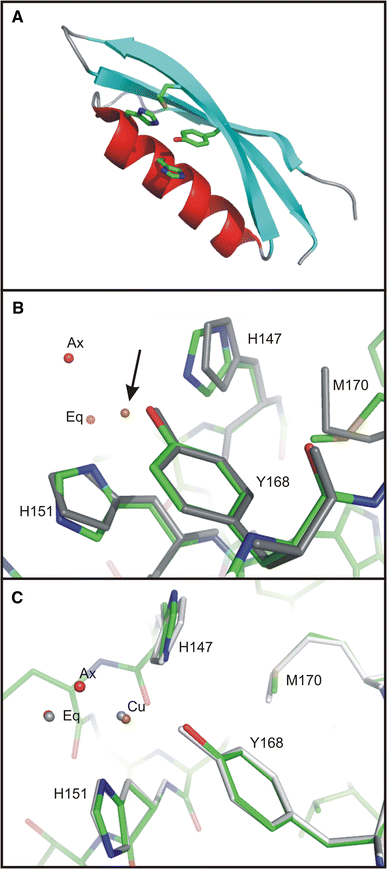
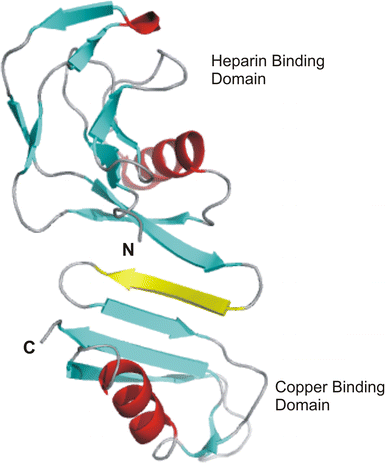
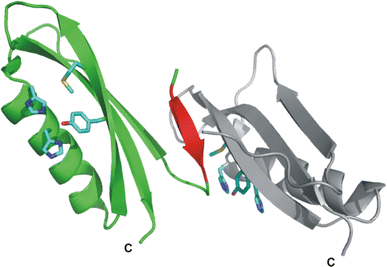
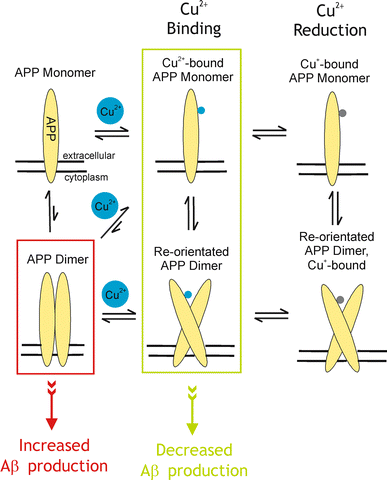
Alzheimer's disease is the fourth biggest killer in developed countries. Amyloid precursor protein (APP) plays a central role in the development of the disease, through the generation of a peptide called A beta by proteolysis of the precursor protein. APP can function as a metalloprotein and modulate copper transport via its extracellular copper binding domain (CuBD). Copper binding to this domain has been shown to reduce A beta levels and hence a molecular understanding of the interaction between metal and protein could lead to the development of novel therapeutics to treat the disease. We have recently determined the three-dimensional structures of apo and copper bound forms of CuBD. The structures provide a mechanism by which CuBD could readily transfer copper ions to other proteins. Importantly, the lack of significant conformational changes to CuBD on copper binding suggests a model in which copper binding affects the dimerisation state of APP leading to reduction in A beta production. We thus predict that disruption of APP dimers may be a novel therapeutic approach to treat Alzheimer's disease.
Figures


Similar articles
-
Structure of the Alzheimer's disease amyloid precursor protein copper binding domain. A regulator of neuronal copper homeostasis.J Biol Chem. 2003 May 9;278(19):17401-7. doi: 10.1074/jbc.M300629200. Epub 2003 Feb 28. J Biol Chem. 2003. PMID: 12611883
-
Structure of Alzheimer's disease amyloid precursor protein copper-binding domain at atomic resolution.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 Oct 1;63(Pt 10):819-24. doi: 10.1107/S1744309107041139. Epub 2007 Sep 19. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007. PMID: 17909280 Free PMC article.
-
Structural studies of the Alzheimer's amyloid precursor protein copper-binding domain reveal how it binds copper ions.J Mol Biol. 2007 Mar 16;367(1):148-61. doi: 10.1016/j.jmb.2006.12.041. Epub 2006 Dec 21. J Mol Biol. 2007. PMID: 17239395
-
Copper brain homeostasis: role of amyloid precursor protein and prion protein.IUBMB Life. 2005 Sep;57(9):645-50. doi: 10.1080/15216540500264620. IUBMB Life. 2005. PMID: 16203684 Review.
-
Copper and Alzheimer's Disease.Adv Neurobiol. 2017;18:199-216. doi: 10.1007/978-3-319-60189-2_10. Adv Neurobiol. 2017. PMID: 28889269 Review.
Cited by
-
Copper Homeostasis in the Model Organism C. elegans.Cells. 2024 Apr 23;13(9):727. doi: 10.3390/cells13090727. Cells. 2024. PMID: 38727263 Free PMC article. Review.
-
Redox signaling and metabolism in Alzheimer's disease.Front Aging Neurosci. 2022 Nov 3;14:1003721. doi: 10.3389/fnagi.2022.1003721. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36408110 Free PMC article. Review.
-
"Pulling the plug" on cellular copper: the role of mitochondria in copper export.Biochim Biophys Acta. 2009 Jan;1793(1):146-53. doi: 10.1016/j.bbamcr.2008.05.002. Epub 2008 May 15. Biochim Biophys Acta. 2009. PMID: 18522804 Free PMC article. Review.
-
The amyloid precursor protein represses expression of acetylcholinesterase in neuronal cell lines.J Biol Chem. 2013 Sep 6;288(36):26039-26051. doi: 10.1074/jbc.M113.461269. Epub 2013 Jul 29. J Biol Chem. 2013. PMID: 23897820 Free PMC article.
-
Alternative Pharmacological Strategies for the Treatment of Alzheimer's Disease: Focus on Neuromodulator Function.Biomedicines. 2022 Nov 28;10(12):3064. doi: 10.3390/biomedicines10123064. Biomedicines. 2022. PMID: 36551821 Free PMC article. Review.
References
-
- Angeletti B, Waldron KJ, Freeman KB, Bawagan H, Hussain I, Miller CCJ, Lau K-F, Tennant ME, Dennison C, Robinson NJ, Dingwall C. BACE1 cytoplasmic domain interacts with the copper chaperone for superoxide dismutase-1 and binds copper. J Biol Chem. 2005;280:17930–17937. doi: 10.1074/jbc.M412034200. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

