Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure
- PMID: 18025469
- PMCID: PMC2141888
- DOI: 10.1073/pnas.0706522104
Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure
Abstract
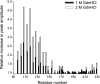
Transmissible spongiform encephalopathies (TSEs) represent a group of fatal neurodegenerative diseases that are associated with conformational conversion of the normally monomeric and alpha-helical prion protein, PrP(C), to the beta-sheet-rich PrP(Sc). This latter conformer is believed to constitute the main component of the infectious TSE agent. In contrast to high-resolution data for the PrP(C) monomer, structures of the pathogenic PrP(Sc) or synthetic PrP(Sc)-like aggregates remain elusive. Here we have used site-directed spin labeling and EPR spectroscopy to probe the molecular architecture of the recombinant PrP amyloid, a misfolded form recently reported to induce transmissible disease in mice overexpressing an N-terminally truncated form of PrP(C). Our data show that, in contrast to earlier, largely theoretical models, the con formational conversion of PrP(C) involves major refolding of the C-terminal alpha-helical region. The core of the amyloid maps to C-terminal residues from approximately 160-220, and these residues form single-molecule layers that stack on top of one another with parallel, in-register alignment of beta-strands. This structural insight has important implications for understanding the molecular basis of prion propagation, as well as hereditary prion diseases, most of which are associated with point mutations in the region found to undergo a refolding to beta-structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Prion protein amyloid formation under native-like conditions involves refolding of the C-terminal alpha-helical domain.J Biol Chem. 2008 Dec 12;283(50):34704-11. doi: 10.1074/jbc.M806701200. Epub 2008 Oct 17. J Biol Chem. 2008. PMID: 18930924 Free PMC article.
-
PrP N-terminal domain triggers PrP(Sc)-like aggregation of Dpl.Biochem Biophys Res Commun. 2008 Jan 18;365(3):478-83. doi: 10.1016/j.bbrc.2007.10.202. Epub 2007 Nov 13. Biochem Biophys Res Commun. 2008. PMID: 17997980
-
Formation of soluble oligomers and amyloid fibrils with physical properties of the scrapie isoform of the prion protein from the C-terminal domain of recombinant murine prion protein mPrP-(121-231).J Biol Chem. 2006 Sep 8;281(36):26121-8. doi: 10.1074/jbc.M605367200. Epub 2006 Jul 13. J Biol Chem. 2006. PMID: 16844683
-
The emerging principles of mammalian prion propagation and transmissibility barriers: Insight from studies in vitro.Acc Chem Res. 2006 Sep;39(9):654-62. doi: 10.1021/ar050226c. Acc Chem Res. 2006. PMID: 16981682 Review.
-
Insights into Mechanisms of Transmission and Pathogenesis from Transgenic Mouse Models of Prion Diseases.Methods Mol Biol. 2017;1658:219-252. doi: 10.1007/978-1-4939-7244-9_16. Methods Mol Biol. 2017. PMID: 28861793 Free PMC article. Review.
Cited by
-
Critical significance of the region between Helix 1 and 2 for efficient dominant-negative inhibition by conversion-incompetent prion protein.PLoS Pathog. 2013;9(6):e1003466. doi: 10.1371/journal.ppat.1003466. Epub 2013 Jun 27. PLoS Pathog. 2013. PMID: 23825952 Free PMC article.
-
Mammalian prions: tolerance to sequence changes-how far?Prion. 2013 Mar-Apr;7(2):131-5. doi: 10.4161/pri.23110. Epub 2012 Dec 11. Prion. 2013. PMID: 23232499 Free PMC article. Review.
-
Generic amyloidogenicity of mammalian prion proteins from species susceptible and resistant to prions.Sci Rep. 2015 May 11;5:10101. doi: 10.1038/srep10101. Sci Rep. 2015. PMID: 25960067 Free PMC article.
-
Highly polar environments catalyze the unfolding of PrP C helix 1.Eur Biophys J. 2010 Jul;39(8):1177-92. doi: 10.1007/s00249-009-0570-6. Epub 2010 Jan 5. Eur Biophys J. 2010. PMID: 20049591
-
Solution structure and dynamics of the I214V mutant of the rabbit prion protein.PLoS One. 2010 Oct 7;5(10):e13273. doi: 10.1371/journal.pone.0013273. PLoS One. 2010. PMID: 20949107 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

