Cloning and characterization of two Lactobacillus casei genes encoding a cystathionine lyase
- PMID: 17993563
- PMCID: PMC2223195
- DOI: 10.1128/AEM.00745-07
Cloning and characterization of two Lactobacillus casei genes encoding a cystathionine lyase
Abstract
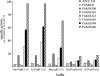
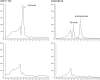
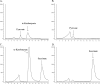
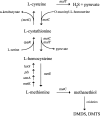
Volatile sulfur compounds are key flavor compounds in several cheese types. To better understand the metabolism of sulfur-containing amino acids, which certainly plays a key role in the release of volatile sulfur compounds, we searched the genome database of Lactobacillus casei ATCC 334 for genes encoding putative homologs of enzymes known to degrade cysteine, cystathionine, and methionine. The search revealed that L. casei possesses two genes that putatively encode a cystathionine beta-lyase (CBL; EC 4.4.1.8). The enzyme has been implicated in the degradation of not only cystathionine but also cysteine and methionine. Recombinant CBL proteins catalyzed the degradation of L-cystathionine, O-succinyl-L-homoserine, L-cysteine, L-serine, and L-methionine to form alpha-keto acid, hydrogen sulfide, or methanethiol. The two enzymes showed notable differences in substrate specificity and pH optimum.
Figures


Similar articles
-
Identification and characterization of a strain-dependent cystathionine beta/gamma-lyase in Lactobacillus casei potentially involved in cysteine biosynthesis.FEMS Microbiol Lett. 2009 Jun;295(1):67-76. doi: 10.1111/j.1574-6968.2009.01580.x. FEMS Microbiol Lett. 2009. PMID: 19473252
-
Properties of recombinant Staphylococcus haemolyticus cystathionine beta-lyase (metC) and its potential role in the generation of volatile thiols in axillary malodor.Chem Biodivers. 2008 Nov;5(11):2372-85. doi: 10.1002/cbdv.200890202. Chem Biodivers. 2008. PMID: 19035565
-
Characterization of C-S lyase from Lactobacillus delbrueckii subsp. bulgaricus ATCC BAA-365 and its potential role in food flavour applications.J Biochem. 2017 Apr 1;161(4):349-360. doi: 10.1093/jb/mvw079. J Biochem. 2017. PMID: 28003427
-
Cloning and characterisation of a cystathionine β/γ-lyase from two Oenococcus oeni oenological strains.Appl Microbiol Biotechnol. 2011 Feb;89(4):1051-60. doi: 10.1007/s00253-010-2911-x. Epub 2010 Oct 5. Appl Microbiol Biotechnol. 2011. PMID: 20922375
-
Sulfur metabolism in bacteria associated with cheese.Antonie Van Leeuwenhoek. 1999 Jul-Nov;76(1-4):247-61. Antonie Van Leeuwenhoek. 1999. PMID: 10532382 Review.
Cited by
-
The Impact of Proteolytic Pork Hydrolysate on Microbial, Flavor and Free Amino Acids Compounds of Yogurt.Korean J Food Sci Anim Resour. 2016;36(4):558-65. doi: 10.5851/kosfa.2016.36.4.558. Epub 2016 Aug 30. Korean J Food Sci Anim Resour. 2016. PMID: 27621698 Free PMC article.
-
Cystathionine gamma-lyase is a component of cystine-mediated oxidative defense in Lactobacillus reuteri BR11.J Bacteriol. 2009 Mar;191(6):1827-37. doi: 10.1128/JB.01553-08. Epub 2009 Jan 5. J Bacteriol. 2009. PMID: 19124577 Free PMC article.
-
Cysteine biosynthesis in Lactobacillus casei: identification and characterization of a serine acetyltransferase.FEMS Microbiol Lett. 2016 Feb;363(4):fnw012. doi: 10.1093/femsle/fnw012. Epub 2016 Jan 19. FEMS Microbiol Lett. 2016. PMID: 26790714 Free PMC article.
-
Microbial pathways in colonic sulfur metabolism and links with health and disease.Front Physiol. 2012 Nov 28;3:448. doi: 10.3389/fphys.2012.00448. eCollection 2012. Front Physiol. 2012. PMID: 23226130 Free PMC article.
-
Underground isoleucine biosynthesis pathways in E. coli.Elife. 2020 Aug 24;9:e54207. doi: 10.7554/eLife.54207. Elife. 2020. PMID: 32831171 Free PMC article.
References
-
- Ardö, Y. 2006. Flavour formation by amino acid catabolism. Biotechnol. Adv. 24:238-242. - PubMed
-
- Aubel, D., J. E. Germond, C. Gilbert, and D. Atlan. 2002. Isolation of the patC gene encoding the cystathionine β-lyase of Lactobacillus delbrueckii subsp. bulgaricus and molecular analysis of inter-strain variability in enzyme biosynthesis. Microbiology 148:2029-2036. - PubMed
-
- Auger, S., M. P. Gomez, A. Danchin, and I. Martin-Verstraete. 2005. The PatB protein of Bacillus subtilis is a C-S-lyase. Biochimie 87:231-238. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

