Protease activity, self interaction, and small interfering RNA binding of the silencing suppressor p1b from cucumber vein yellowing ipomovirus
- PMID: 17989179
- PMCID: PMC2224578
- DOI: 10.1128/JVI.01664-07
Protease activity, self interaction, and small interfering RNA binding of the silencing suppressor p1b from cucumber vein yellowing ipomovirus
Abstract
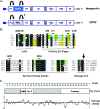
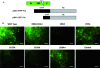
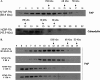
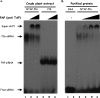
The RNA silencing pathway mediated by small interfering RNAs (siRNAs) plays an important antiviral role in eukaryotes. To counteract this defense barrier, a large number of plant viruses express proteins with RNA silencing suppression activity. Recently, it was reported that the ipomovirus Cucumber vein yellowing virus (CVYV), which lacks the typical silencing suppressor of members of the family Potyviridae, i.e., HCPro, has a duplicated P1 coding sequence and that the downstream P1 copy, named P1b, has silencing suppression activity. In this study, we provide experimental evidence that P1b is a serine protease that self-cleaves at its C terminus but that its proteolytic activity is not essential for silencing suppression. In contrast, a putative zinc finger and a conserved basic motif in the N-terminal region of the protein are required for efficient silencing suppression. In vitro gel filtration-fast protein liquid chromatography and in vivo bimolecular fluorescence complementation assays showed that P1b binds itself to form oligomeric structures and that the zinc finger-like motif is essential for the self interaction. Moreover, we observed that CVYV P1b forms complexes with synthetic siRNAs, and this ability correlated with both silencing suppression activity and enhancement of Potato virus X pathogenicity in a mutational analysis. Together, these results suggest that CVYV P1b resembles potyviral HCPro and other viral proteins in interfering RNA silencing by preventing siRNA loading into the RNA-induced silencing complex.
Figures




Similar articles
-
RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro.J Virol. 2006 Oct;80(20):10055-63. doi: 10.1128/JVI.00985-06. J Virol. 2006. PMID: 17005683 Free PMC article.
-
The specific binding to 21-nt double-stranded RNAs is crucial for the anti-silencing activity of Cucumber vein yellowing virus P1b and perturbs endogenous small RNA populations.RNA. 2011 Jun;17(6):1148-58. doi: 10.1261/rna.2510611. Epub 2011 Apr 29. RNA. 2011. PMID: 21531919 Free PMC article.
-
The Cucumber vein yellowing virus silencing suppressor P1b can functionally replace HCPro in Plum pox virus infection in a host-specific manner.Mol Plant Microbe Interact. 2012 Feb;25(2):151-64. doi: 10.1094/MPMI-08-11-0216. Mol Plant Microbe Interact. 2012. PMID: 21970691
-
P1 peptidase--a mysterious protein of family Potyviridae.J Biosci. 2011 Mar;36(1):189-200. doi: 10.1007/s12038-011-9020-6. J Biosci. 2011. PMID: 21451259 Review.
-
The HCPro from the Potyviridae family: an enviable multitasking Helper Component that every virus would like to have.Mol Plant Pathol. 2018 Mar;19(3):744-763. doi: 10.1111/mpp.12553. Epub 2017 May 26. Mol Plant Pathol. 2018. PMID: 28371183 Free PMC article. Review.
Cited by
-
Plant Viral Proteases: Beyond the Role of Peptide Cutters.Front Plant Sci. 2018 May 17;9:666. doi: 10.3389/fpls.2018.00666. eCollection 2018. Front Plant Sci. 2018. PMID: 29868107 Free PMC article. Review.
-
Identification of the amino acid residues and domains in the cysteine-rich protein of Chinese wheat mosaic virus that are important for RNA silencing suppression and subcellular localization.Mol Plant Pathol. 2013 Apr;14(3):265-78. doi: 10.1111/mpp.12002. Mol Plant Pathol. 2013. PMID: 23458485 Free PMC article.
-
Molecular characterization of Indian sugarcane streak mosaic virus isolate.Virus Genes. 2013 Feb;46(1):186-9. doi: 10.1007/s11262-012-0827-5. Epub 2012 Sep 26. Virus Genes. 2013. PMID: 23011777
-
Bymovirus reverse genetics: requirements for RNA2-encoded proteins in systemic infection.Mol Plant Pathol. 2010 May;11(3):383-94. doi: 10.1111/j.1364-3703.2010.00613.x. Mol Plant Pathol. 2010. PMID: 20447286 Free PMC article.
-
Complete genomic sequence analyses of Turnip mosaic virus basal-BR isolates from China.Virus Genes. 2009 Jun;38(3):421-8. doi: 10.1007/s11262-009-0335-4. Epub 2009 Feb 24. Virus Genes. 2009. PMID: 19238532
References
-
- Alamillo, J. M., W. Monger, I. Sola, B. Garcia, Y. Perrin, M. Bestagno, O. R. Burrone, P. Sabella, J. Plana-Duran, L. Enjuanes, G. P. Lomonossoff, and J. A. Garcia. 2006. Use of virus vectors for the expression in plants of active full-length and single chain anti-coronavirus antibodies. Biotechnol. J. 11103-1111. - PMC - PubMed
-
- Ambros, V., and X. M. Chen. 2007. The regulation of genes and genomes by small RNAs. Development 1341635-1641. - PubMed
-
- Baulcombe, D. 2002. RNA silencing. Curr. Biol. 12R82-R84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

