Nitric oxide destabilizes Pias3 and regulates sumoylation
- PMID: 17987106
- PMCID: PMC2064872
- DOI: 10.1371/journal.pone.0001085
Nitric oxide destabilizes Pias3 and regulates sumoylation
Abstract
Small ubiquitin-related protein modifiers (SUMO) modification is an important mechanism for posttranslational regulation of protein function. However, it is largely unknown how the sumoylation pathway is regulated. Here, we report that nitric oxide (NO) causes global hyposumoylation in mammalian cells. Both SUMO E2 conjugating enzyme Ubc9 and E3 ligase protein inhibitor of activated STAT3 (Pias3) were targets for S-nitrosation. S-nitrosation did not interfere with the SUMO conjugating activity of Ubc9, but promoted Pias3 degradation by facilitating its interaction with tripartite motif-containing 32 (Trim32), a ubiquitin E3 ligase. On the one hand, NO promoted Trim32-mediated Pias3 ubiquitination. On the other hand, NO enhanced the stimulatory effect of Pias3 on Trim32 autoubiquitination. The residue Cys459 of Pias3 was identified as a target site for S-nitrosation. Mutation of Cys459 abolished the stimulatory effect of NO on the Pias3-Trim32 interaction, indicating a requirement of S-nitrosation at Cys459 for positive regulation of the Pias3-Trim32 interplay. This study reveals a novel crosstalk between S-nitrosation, ubiquitination, and sumoylation, which may be crucial for NO-related physiological and pathological processes.
Conflict of interest statement
Figures
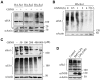

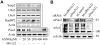
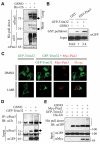
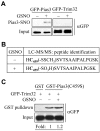
Similar articles
-
Activity-Induced SUMOylation of Neuronal Nitric Oxide Synthase Is Associated with Plasticity of Synaptic Transmission and Extracellular Signal-Regulated Kinase 1/2 Signaling.Antioxid Redox Signal. 2020 Jan 1;32(1):18-34. doi: 10.1089/ars.2018.7669. Antioxid Redox Signal. 2020. PMID: 31642335
-
Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity.Mol Endocrinol. 2005 Nov;19(11):2671-84. doi: 10.1210/me.2005-0042. Epub 2005 Jun 16. Mol Endocrinol. 2005. PMID: 15961505
-
The SUMO-E3 ligase PIAS3 targets pyruvate kinase M2.J Cell Biochem. 2009 May 15;107(2):293-302. doi: 10.1002/jcb.22125. J Cell Biochem. 2009. PMID: 19308990
-
Novel initiation genes in squamous cell carcinomagenesis: a role for substrate-specific ubiquitylation in the control of cell survival.Mol Carcinog. 2007 Aug;46(8):585-90. doi: 10.1002/mc.20344. Mol Carcinog. 2007. PMID: 17626251 Review.
-
SUMO pathway components as possible cancer biomarkers.Future Oncol. 2015;11(11):1599-610. doi: 10.2217/fon.15.41. Future Oncol. 2015. PMID: 26043214 Review.
Cited by
-
Precision Redox: The Key for Antioxidant Pharmacology.Antioxid Redox Signal. 2021 May 10;34(14):1069-1082. doi: 10.1089/ars.2020.8212. Epub 2020 Dec 2. Antioxid Redox Signal. 2021. PMID: 33270507 Free PMC article. Review.
-
Nuclear Smad6 promotes gliomagenesis by negatively regulating PIAS3-mediated STAT3 inhibition.Nat Commun. 2018 Jun 27;9(1):2504. doi: 10.1038/s41467-018-04936-9. Nat Commun. 2018. PMID: 29950561 Free PMC article.
-
Ubiquitin-Conjugating Enzyme 9 Phosphorylation as a Novel Mechanism for Potentiation of the Inflammatory Response.Am J Pathol. 2016 Sep;186(9):2326-36. doi: 10.1016/j.ajpath.2016.05.007. Am J Pathol. 2016. PMID: 27561301 Free PMC article.
-
Lipin proteins form homo- and hetero-oligomers.Biochem J. 2010 Nov 15;432(1):65-76. doi: 10.1042/BJ20100584. Biochem J. 2010. PMID: 20735359 Free PMC article.
-
Gating neural development and aging via nuclear pores.Cell Res. 2012 Aug;22(8):1212-4. doi: 10.1038/cr.2012.35. Epub 2012 Mar 13. Cell Res. 2012. PMID: 22410792 Free PMC article.
References
-
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. - PubMed
-
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. - PubMed
-
- Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. - PubMed
-
- Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16:549–561. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

