Functional coupling of last-intron splicing and 3'-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage
- PMID: 17967872
- PMCID: PMC2223410
- DOI: 10.1128/MCB.01410-07
Functional coupling of last-intron splicing and 3'-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage
Abstract
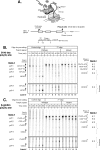
We have developed an in vitro transcription system, using HeLa nuclear extract, that supports not only efficient splicing of a multiexon transcript but also efficient cleavage and polyadenylation. In this system, both last-intron splicing and cleavage/polyadenylation are functionally coupled to transcription via the tether of nascent RNA that extends from the terminal exon to the transcribing polymerase downstream. Communication between the 3' splice site and the poly(A) site across the terminal exon is established within minutes of their transcription, and multiple steps leading up to 3'-end processing of this exon can be distinguished. First, the 3' splice site establishes connections to enhance 3'-end processing, while the nascent 3'-end processing apparatus makes reciprocal functional connections to enhance splicing. Then, commitment to poly(A) site cleavage itself occurs and the connections of the 3'-end processing apparatus to the transcribing polymerase are strengthened. Finally, the chemical steps in the processing of the terminal exon take place, beginning with poly(A) site cleavage, continuing with polyadenylation of the 3' end, and then finishing with splicing of the last intron.
Figures




Similar articles
-
An active role for splicing in 3'-end formation.Wiley Interdiscip Rev RNA. 2011 Jul-Aug;2(4):459-70. doi: 10.1002/wrna.68. Epub 2010 Dec 16. Wiley Interdiscip Rev RNA. 2011. PMID: 21957037 Review.
-
An intron enhancer recognized by splicing factors activates polyadenylation.Genes Dev. 1996 Jan 15;10(2):208-19. doi: 10.1101/gad.10.2.208. Genes Dev. 1996. PMID: 8566754
-
Upstream introns influence the efficiency of final intron removal and RNA 3'-end formation.Genes Dev. 1994 Feb 1;8(3):363-75. doi: 10.1101/gad.8.3.363. Genes Dev. 1994. PMID: 7906237
-
Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns.Genes Dev. 1991 Nov;5(11):2086-95. doi: 10.1101/gad.5.11.2086. Genes Dev. 1991. PMID: 1657710
-
A history of poly A sequences: from formation to factors to function.Prog Nucleic Acid Res Mol Biol. 2002;71:285-389. doi: 10.1016/s0079-6603(02)71046-5. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12102557 Review.
Cited by
-
U1snRNP-mediated suppression of polyadenylation in conjunction with the RNA structure controls poly (A) site selection in foamy viruses.Retrovirology. 2013 May 29;10:55. doi: 10.1186/1742-4690-10-55. Retrovirology. 2013. PMID: 23718736 Free PMC article.
-
Poly(A) signal-dependent degradation of unprocessed nascent transcripts accompanies poly(A) signal-dependent transcriptional pausing in vitro.RNA. 2010 Jan;16(1):197-210. doi: 10.1261/rna.1622010. Epub 2009 Nov 19. RNA. 2010. PMID: 19926725 Free PMC article.
-
Association of mitochondrial DNA polymerase γ gene POLG1 polymorphisms with parkinsonism in Chinese populations.PLoS One. 2012;7(12):e50086. doi: 10.1371/journal.pone.0050086. Epub 2012 Dec 10. PLoS One. 2012. PMID: 23251356 Free PMC article.
-
Plasma cell formation, secretion, and persistence: the short and the long of it.Crit Rev Immunol. 2014;34(6):481-99. doi: 10.1615/critrevimmunol.2014012168. Crit Rev Immunol. 2014. PMID: 25597311 Free PMC article. Review.
-
Juxtaposition of two distant, serine-arginine-rich protein-binding elements is required for optimal polyadenylation in Rous sarcoma virus.J Virol. 2011 Nov;85(21):11351-60. doi: 10.1128/JVI.00721-11. Epub 2011 Aug 17. J Virol. 2011. PMID: 21849435 Free PMC article.
References
-
- Adamson, T. E., D. C. Shutt, and D. H. Price. 2005. Functional coupling of cleavage and polyadenylation with transcription of mRNA. J. Biol. Chem. 28032262-32271. - PubMed
-
- Bentley, D. L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17251-256. - PubMed
-
- Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 2702411-2414. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
