Crystal structure of the NS3 protease-helicase from dengue virus
- PMID: 17942558
- PMCID: PMC2224403
- DOI: 10.1128/JVI.01788-07
Crystal structure of the NS3 protease-helicase from dengue virus
Abstract
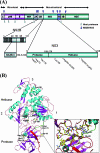
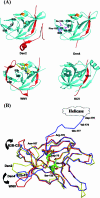
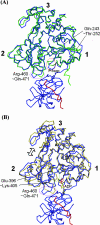
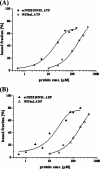
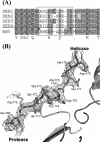
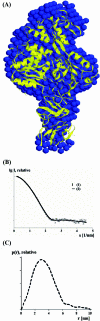
Several flaviviruses are important human pathogens, including dengue virus, a disease against which neither a vaccine nor specific antiviral therapies currently exist. During infection, the flavivirus RNA genome is translated into a polyprotein, which is cleaved into several components. Nonstructural protein 3 (NS3) carries out enzymatic reactions essential for viral replication, including proteolysis of the polyprotein through its serine protease N-terminal domain, with a segment of 40 residues from the NS2B protein acting as a cofactor. The ATPase/helicase domain is located at the C terminus of NS3. Atomic structures are available for these domains separately, but a molecular view of the full-length flavivirus NS3 polypeptide is still lacking. We report a crystallographic structure of a complete NS3 molecule fused to 18 residues of the NS2B cofactor at a resolution of 3.15 A. The relative orientation between the protease and helicase domains is drastically different than the single-chain NS3-NS4A molecule from hepatitis C virus, which was caught in the act of cis cleavage at the NS3-NS4A junction. Here, the protease domain sits beneath the ATP binding site, giving the molecule an elongated shape. The domain arrangement found in the crystal structure fits nicely into an envelope determined ab initio using small-angle X-ray scattering experiments in solution, suggesting a stable molecular conformation. We propose that a basic patch located at the surface of the protease domain increases the affinity for nucleotides and could also participate in RNA binding, explaining the higher unwinding activity of the full-length enzyme compared to that of the isolated helicase domain.
Figures

Similar articles
-
Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase.Structure. 1999 Nov 15;7(11):1353-63. doi: 10.1016/s0969-2126(00)80025-8. Structure. 1999. PMID: 10574797
-
Dynamic Interactions of Post Cleaved NS2B Cofactor and NS3 Protease Identified by Integrative Structural Approaches.Viruses. 2022 Jun 30;14(7):1440. doi: 10.3390/v14071440. Viruses. 2022. PMID: 35891424 Free PMC article.
-
Uncoupling of Protease trans-Cleavage and Helicase Activities in Pestivirus NS3.J Virol. 2017 Oct 13;91(21):e01094-17. doi: 10.1128/JVI.01094-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28835495 Free PMC article.
-
Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target.Antiviral Res. 2008 Nov;80(2):94-101. doi: 10.1016/j.antiviral.2008.07.001. Epub 2008 Jul 30. Antiviral Res. 2008. PMID: 18674567 Review.
-
The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development.Antiviral Res. 2015 Jun;118:148-58. doi: 10.1016/j.antiviral.2015.03.014. Epub 2015 Apr 2. Antiviral Res. 2015. PMID: 25842996 Review.
Cited by
-
X-ray structure of the pestivirus NS3 helicase and its conformation in solution.J Virol. 2015 Apr;89(8):4356-71. doi: 10.1128/JVI.03165-14. Epub 2015 Feb 4. J Virol. 2015. PMID: 25653438 Free PMC article.
-
NLRC5 restricts dengue virus infection by promoting the autophagic degradation of viral NS3 through E3 ligase CUL2 (cullin 2).Autophagy. 2023 Apr;19(4):1332-1347. doi: 10.1080/15548627.2022.2126614. Epub 2022 Sep 29. Autophagy. 2023. PMID: 36126167 Free PMC article.
-
The core protein of classical Swine Fever virus is dispensable for virus propagation in vitro.PLoS Pathog. 2012;8(3):e1002598. doi: 10.1371/journal.ppat.1002598. Epub 2012 Mar 22. PLoS Pathog. 2012. PMID: 22457622 Free PMC article.
-
Proteases from dengue, West Nile and Zika viruses as drug targets.Biophys Rev. 2019 Apr;11(2):157-165. doi: 10.1007/s12551-019-00508-3. Epub 2019 Feb 26. Biophys Rev. 2019. PMID: 30806881 Free PMC article. Review.
-
Thermodynamic and mechanistic analysis of the functional properties of dengue virus NS3 helicase.Biophys Rev. 2023 Aug 17;15(4):591-600. doi: 10.1007/s12551-023-01101-5. eCollection 2023 Aug. Biophys Rev. 2023. PMID: 37681085 Free PMC article. Review.
References
-
- Arias, C. F., F. Preugschat, and J. H. Strauss. 1993. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology 193888-899. - PubMed
-
- Armbrüster, A., D. I. Svergun, U. Coskun, S. Juliano, S. M. Bailer, and G. Grüber. 2004. Structural analysis of the stalk subunit Vma5p of the yeast V-ATPase in solution. FEBS Lett. 570119-125. - PubMed
-
- Bera, A. K., R. J. Kuhn, and J. L. Smith. 2007. Functional characterization of cis and trans activity of the flavivirus NS2B-NS3 protease. J. Biol. Chem. 28212883-12892. - PubMed
-
- Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44649-688. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

