The EGF receptor is required for efficient liver regeneration
- PMID: 17940036
- PMCID: PMC2040457
- DOI: 10.1073/pnas.0704126104
The EGF receptor is required for efficient liver regeneration
Erratum in
- Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19656
Abstract
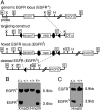
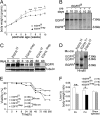
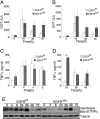
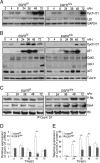
Mice lacking the EGF receptor (EGFR) die between midgestation and postnatal day 20 with various defects in neural and epithelial organs. Here, we generated mice carrying a floxed EGFR allele to inactivate the EGFR in fetal and adult liver. Perinatal deletion of EGFR in hepatocytes resulted in decreased body weight, whereas deletion in the adult liver did not affect body mass. Although liver function was not affected, after partial hepatectomy mice lacking EGFR in the liver showed increased mortality accompanied by increased levels of serum transaminases indicating liver damage. Liver regeneration was delayed in the mutants because of reduced hepatocyte proliferation. Analysis of cell cycle progression in EGFR-deficient livers indicated a defective G(1)-S phase entry with delayed transcriptional activation and reduced protein expression of cyclin D1 followed by reduced cdk2 and cdk1 expression. Impaired liver regeneration was accompanied by compensatory up-regulation of TNFalpha in the serum and prolonged activation of c-Jun. Moreover, p38alpha and NF-kappaB activation was reduced in regenerating mutant livers, indicating an impaired stress response after hepatectomy. Our studies demonstrate that EGFR is a critical regulator of hepatocyte proliferation in the initial phases of liver regeneration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver.EMBO J. 2002 Apr 2;21(7):1782-90. doi: 10.1093/emboj/21.7.1782. EMBO J. 2002. PMID: 11927562 Free PMC article.
-
ZBTB20 regulates EGFR expression and hepatocyte proliferation in mouse liver regeneration.Cell Death Dis. 2018 May 1;9(5):462. doi: 10.1038/s41419-018-0514-0. Cell Death Dis. 2018. PMID: 29700307 Free PMC article.
-
Deregulation of growth factor, circadian clock, and cell cycle signaling in regenerating hepatocyte RXRalpha-deficient mouse livers.Am J Pathol. 2010 Feb;176(2):733-43. doi: 10.2353/ajpath.2010.090524. Epub 2009 Dec 24. Am J Pathol. 2010. PMID: 20035057 Free PMC article.
-
Hepatocyte-specific inhibitor-of-kappaB-kinase deletion triggers the innate immune response and promotes earlier cell proliferation during liver regeneration.Hepatology. 2008 Jun;47(6):2036-50. doi: 10.1002/hep.22264. Hepatology. 2008. PMID: 18393321
-
Signal transduction during liver regeneration.J Gastroenterol Hepatol. 1998 Sep;13 Suppl:S93-5. J Gastroenterol Hepatol. 1998. PMID: 9792040 Review.
Cited by
-
A disintegrin and metalloproteinase 17 regulates TNF and TNFR1 levels in inflammation and liver regeneration in mice.Am J Physiol Gastrointest Liver Physiol. 2013 Jul 1;305(1):G25-34. doi: 10.1152/ajpgi.00326.2012. Epub 2013 May 2. Am J Physiol Gastrointest Liver Physiol. 2013. PMID: 23639813 Free PMC article.
-
FAK deletion accelerates liver regeneration after two-thirds partial hepatectomy.Sci Rep. 2016 Sep 28;6:34316. doi: 10.1038/srep34316. Sci Rep. 2016. PMID: 27677358 Free PMC article.
-
Epidermal growth factor regulates hematopoietic regeneration after radiation injury.Nat Med. 2013 Mar;19(3):295-304. doi: 10.1038/nm.3070. Epub 2013 Feb 3. Nat Med. 2013. PMID: 23377280 Free PMC article.
-
Gasdermin D-mediated pyroptosis suppresses liver regeneration after 70% partial hepatectomy.Hepatol Commun. 2022 Sep;6(9):2340-2353. doi: 10.1002/hep4.1973. Epub 2022 May 4. Hepatol Commun. 2022. PMID: 35509206 Free PMC article.
-
Blockade of the P2Y2 Receptor Attenuates Alcoholic Liver Inflammation by Targeting the EGFR-ERK1/2 Signaling Pathway.Drug Des Devel Ther. 2022 Apr 13;16:1107-1120. doi: 10.2147/DDDT.S346376. eCollection 2022. Drug Des Devel Ther. 2022. PMID: 35444406 Free PMC article.
References
-
- Carver RS, Stevenson MC, Scheving LA, Russell WE. Gastroenterology. 2002;123:2017–2027. - PubMed
-
- Yarden Y, Sliwkowski MX. Nat Rev Mol Cell Biol. 2001;2:127–137. - PubMed
-
- Schlessinger J. Cell. 2000;103:211–225. - PubMed
-
- Schlessinger J. Cell. 2002;110:669–672. - PubMed
-
- Fausto N, Campbell JS, Riehle KJ. Hepatology. 2006;43:S45–S53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

