HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription
- PMID: 17937499
- PMCID: PMC2014796
- DOI: 10.1371/journal.ppat.0030146
HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription
Abstract
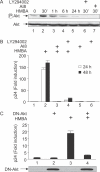
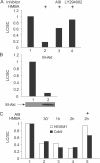
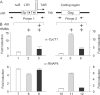
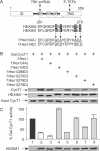
Hexamethylene bisacetamide (HMBA) is a potent inducer of cell differentiation and HIV production in chronically infected cells. However, its mechanism of action remains poorly defined. In this study, we demonstrate that HMBA activates transiently the PI3K/Akt pathway, which leads to the phosphorylation of HEXIM1 and the subsequent release of active positive transcription elongation factor b (P-TEFb) from its transcriptionally inactive complex with HEXIM1 and 7SK small nuclear RNA (snRNA). As a result, P-TEFb is recruited to the HIV promoter to stimulate transcription elongation and viral production. Despite the continuous presence of HMBA, the released P-TEFb reassembles rapidly with 7SK snRNA and HEXIM1. In contrast, a mutant HEXIM1 protein that cannot be phosphorylated and released from P-TEFb and 7SK snRNA via the PI3K/Akt pathway antagonizes this HMBA-mediated induction of viral production. Thus, our studies reveal how HIV transcription is induced by HMBA and suggest how modifications in the equilibrium between active and inactive P-TEFb could contribute to cell differentiation.
Conflict of interest statement
Figures

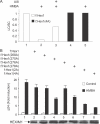
Similar articles
-
Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein.J Biol Chem. 2012 Oct 19;287(43):36609-16. doi: 10.1074/jbc.M112.410746. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952229 Free PMC article.
-
Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription.Nucleic Acids Res. 2007;35(6):2003-12. doi: 10.1093/nar/gkm063. Epub 2007 Mar 6. Nucleic Acids Res. 2007. PMID: 17341462 Free PMC article.
-
Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation.Mol Cell Biol. 2006 Oct;26(19):7068-76. doi: 10.1128/MCB.00778-06. Mol Cell Biol. 2006. PMID: 16980611 Free PMC article.
-
Brd4 and HEXIM1: multiple roles in P-TEFb regulation and cancer.Biomed Res Int. 2014;2014:232870. doi: 10.1155/2014/232870. Epub 2014 Jan 29. Biomed Res Int. 2014. PMID: 24592384 Free PMC article. Review.
-
HEXIM1 and the control of transcription elongation: from cancer and inflammation to AIDS and cardiac hypertrophy.Cell Cycle. 2007 Aug 1;6(15):1856-63. doi: 10.4161/cc.6.15.4556. Epub 2007 Jun 6. Cell Cycle. 2007. PMID: 17671421 Review.
Cited by
-
Stable Phenotypic Changes of the Host T Cells Are Essential to the Long-Term Stability of Latent HIV-1 Infection.J Virol. 2015 Jul;89(13):6656-72. doi: 10.1128/JVI.00571-15. Epub 2015 Apr 15. J Virol. 2015. PMID: 25878110 Free PMC article.
-
Isolation and functional characterization of P-TEFb-associated factors that control general and HIV-1 transcriptional elongation.Methods. 2011 Jan;53(1):85-90. doi: 10.1016/j.ymeth.2010.04.005. Epub 2010 Apr 10. Methods. 2011. PMID: 20385240 Free PMC article.
-
Alternate NF-κB-Independent Signaling Reactivation of Latent HIV-1 Provirus.J Virol. 2019 Aug 28;93(18):e00495-19. doi: 10.1128/JVI.00495-19. Print 2019 Sep 15. J Virol. 2019. PMID: 31243131 Free PMC article.
-
Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells.EMBO J. 2009 May 20;28(10):1407-17. doi: 10.1038/emboj.2009.99. Epub 2009 Apr 23. EMBO J. 2009. PMID: 19387490 Free PMC article.
-
Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2.J Virol. 2011 Sep;85(17):9078-89. doi: 10.1128/JVI.00836-11. Epub 2011 Jun 29. J Virol. 2011. PMID: 21715480 Free PMC article.
References
-
- Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF. The multifactorial nature of HIV-1 latency. Trends Mol Med. 2004;10:525–531. - PubMed
-
- Chun TW, Engel D, Mizell SB, Hallahan CW, Fischette M, et al. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat Med. 1999;5:651–655. - PubMed
-
- Stellbrink HJ, van Lunzen J, Westby M, O'Sullivan E, Schneider C, et al. Effects of interleukin-2 plus highly active antiretroviral therapy on HIV-1 replication and proviral DNA (COSMIC trial) AIDS. 2002;16:1479–1487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

