Dynamics of human replication factors in the elongation phase of DNA replication
- PMID: 17932049
- PMCID: PMC2175312
- DOI: 10.1093/nar/gkm822
Dynamics of human replication factors in the elongation phase of DNA replication
Abstract
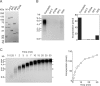
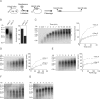
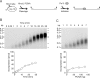
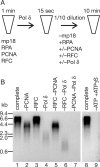
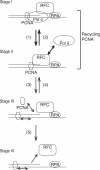
In eukaryotic cells, DNA replication is carried out by coordinated actions of many proteins, including DNA polymerase delta (pol delta), replication factor C (RFC), proliferating cell nuclear antigen (PCNA) and replication protein A. Here we describe dynamic properties of these proteins in the elongation step on a single-stranded M13 template, providing evidence that pol delta has a distributive nature over the 7 kb of the M13 template, repeating a frequent dissociation-association cycle at growing 3'-hydroxyl ends. Some PCNA could remain at the primer terminus during this cycle, while the remainder slides out of the primer terminus or is unloaded once pol delta has dissociated. RFC remains around the primer terminus through the elongation phase, and could probably hold PCNA from which pol delta has detached, or reload PCNA from solution to restart DNA synthesis. Furthermore, we suggest that a subunit of pol delta, POLD3, plays a crucial role in the efficient recycling of PCNA during dissociation-association cycles of pol delta. Based on these observations, we propose a model for dynamic processes in elongation complexes.
Figures



Similar articles
-
Replication factor C disengages from proliferating cell nuclear antigen (PCNA) upon sliding clamp formation, and PCNA itself tethers DNA polymerase delta to DNA.J Biol Chem. 1998 Nov 27;273(48):31992-9. doi: 10.1074/jbc.273.48.31992. J Biol Chem. 1998. PMID: 9822671
-
Functional sites of human PCNA which interact with p21 (Cip1/Waf1), DNA polymerase delta and replication factor C.Genes Cells. 1998 Jun;3(6):357-69. doi: 10.1046/j.1365-2443.1998.00199.x. Genes Cells. 1998. PMID: 9734782
-
Replication protein A dynamically re-organizes on primer/template junctions to permit DNA polymerase δ holoenzyme assembly and initiation of DNA synthesis.Nucleic Acids Res. 2024 Jul 22;52(13):7650-7664. doi: 10.1093/nar/gkae475. Nucleic Acids Res. 2024. PMID: 38842913 Free PMC article.
-
DNA polymerase delta, an essential enzyme for DNA transactions.Biol Chem. 1997 May;378(5):345-62. doi: 10.1515/bchm.1997.378.5.345. Biol Chem. 1997. PMID: 9191022 Review.
-
Functions of Multiple Clamp and Clamp-Loader Complexes in Eukaryotic DNA Replication.Adv Exp Med Biol. 2017;1042:135-162. doi: 10.1007/978-981-10-6955-0_7. Adv Exp Med Biol. 2017. PMID: 29357057 Review.
Cited by
-
Stability of the human polymerase δ holoenzyme and its implications in lagging strand DNA synthesis.Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):E1777-86. doi: 10.1073/pnas.1523653113. Epub 2016 Mar 14. Proc Natl Acad Sci U S A. 2016. PMID: 26976599 Free PMC article.
-
Human replication factor C stimulates flap endonuclease 1.J Biol Chem. 2009 Apr 17;284(16):10387-99. doi: 10.1074/jbc.M808893200. Epub 2009 Feb 9. J Biol Chem. 2009. PMID: 19208620 Free PMC article.
-
Rad51 and Rad54 promote noncrossover recombination between centromere repeats on the same chromatid to prevent isochromosome formation.Nucleic Acids Res. 2016 Dec 15;44(22):10744-10757. doi: 10.1093/nar/gkw874. Epub 2016 Oct 3. Nucleic Acids Res. 2016. PMID: 27697832 Free PMC article.
-
Functions of alternative replication protein A in initiation and elongation.Biochemistry. 2010 Jul 20;49(28):5919-28. doi: 10.1021/bi100380n. Biochemistry. 2010. PMID: 20545304 Free PMC article.
-
Determination of the biochemical properties of full-length human PIF1 ATPase.Prion. 2013 Jul-Aug;7(4):341-7. doi: 10.4161/pri.26022. Epub 2013 Aug 7. Prion. 2013. PMID: 23924759 Free PMC article.
References
-
- Hübscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. - PubMed
-
- Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. - PubMed
-
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998;67:721–751. - PubMed
-
- Stillman B. Initiation of eukaryotic DNA replication in vitro. Annu. Rev. Cell. Biol. 1989;5:197–245. - PubMed
-
- MacNeill SA, Baldacci G, Burgers PM, Hübscher U. A unified nomenclature for the subunits of eukaryotic DNA polymerase δ. Trends Biochem. Sci. 2001;26:16–17. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

