Structure, dynamics, and evolution of centromeric nucleosomes
- PMID: 17893333
- PMCID: PMC1993840
- DOI: 10.1073/pnas.0707648104
Structure, dynamics, and evolution of centromeric nucleosomes
Abstract
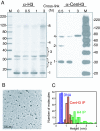
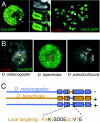
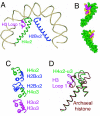
Centromeres are defining features of eukaryotic chromosomes, providing sites of attachment for segregation during mitosis and meiosis. The fundamental unit of centromere structure is the centromeric nucleosome, which differs from the conventional nucleosome by the presence of a centromere-specific histone variant (CenH3) in place of canonical H3. We have shown that the CenH3 nucleosome core found in interphase Drosophila cells is a heterotypic tetramer, a "hemisome" consisting of one molecule each of CenH3, H4, H2A, and H2B, rather than the octamer of canonical histones that is found in bulk nucleosomes. The surprising discovery of hemisomes at centromeres calls for a reevaluation of evidence that has long been interpreted in terms of a more conventional nucleosome. We describe how the hemisome structure of centromeric nucleosomes can account for enigmatic properties of centromeres, including kinetochore accessibility, epigenetic inheritance, rapid turnover of misincorporated CenH3, and transcriptional quiescence of pericentric heterochromatin. Structural differences mediated by loop 1 are proposed to account for the formation of stable tetramers containing CenH3 rather than stable octamers containing H3. Asymmetric CenH3 hemisomes might interrupt the global condensation of octameric H3 arrays and present an asymmetric surface for kinetochore formation. We suggest that this simple mechanism for differentiation between centromeric and packaging nucleosomes evolved from an archaea-like ancestor at the dawn of eukaryotic evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells.PLoS Biol. 2007 Aug;5(8):e218. doi: 10.1371/journal.pbio.0050218. PLoS Biol. 2007. PMID: 17676993 Free PMC article.
-
The unconventional structure of centromeric nucleosomes.Chromosoma. 2012 Aug;121(4):341-52. doi: 10.1007/s00412-012-0372-y. Epub 2012 May 3. Chromosoma. 2012. PMID: 22552438 Free PMC article. Review.
-
Tetrameric organization of vertebrate centromeric nucleosomes.Proc Natl Acad Sci U S A. 2010 Nov 23;107(47):20317-22. doi: 10.1073/pnas.1009563107. Epub 2010 Nov 8. Proc Natl Acad Sci U S A. 2010. PMID: 21059934 Free PMC article.
-
Chaperone-mediated assembly of centromeric chromatin in vitro.Proc Natl Acad Sci U S A. 2006 Apr 18;103(16):6172-7. doi: 10.1073/pnas.0601686103. Epub 2006 Apr 6. Proc Natl Acad Sci U S A. 2006. PMID: 16601098 Free PMC article.
-
Epigenetic inheritance of centromeres.Cold Spring Harb Symp Quant Biol. 2010;75:51-60. doi: 10.1101/sqb.2010.75.001. Epub 2010 Nov 3. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21047902 Review.
Cited by
-
Neocentromeres form efficiently at multiple possible loci in Candida albicans.PLoS Genet. 2009 Mar;5(3):e1000400. doi: 10.1371/journal.pgen.1000400. Epub 2009 Mar 6. PLoS Genet. 2009. PMID: 19266018 Free PMC article.
-
Stable complex formation of CENP-B with the CENP-A nucleosome.Nucleic Acids Res. 2015 May 26;43(10):4909-22. doi: 10.1093/nar/gkv405. Epub 2015 Apr 27. Nucleic Acids Res. 2015. PMID: 25916850 Free PMC article.
-
Crystal structure of the human centromeric nucleosome containing CENP-A.Nature. 2011 Jul 10;476(7359):232-5. doi: 10.1038/nature10258. Nature. 2011. PMID: 21743476
-
The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere.Proc Natl Acad Sci U S A. 2011 Aug 2;108(31):12687-92. doi: 10.1073/pnas.1104978108. Epub 2011 Jul 18. Proc Natl Acad Sci U S A. 2011. PMID: 21768332 Free PMC article.
-
Sequence information encoded in DNA that may influence long-range chromatin structure correlates with human chromosome functions.PLoS One. 2008 Jul 9;3(7):e2643. doi: 10.1371/journal.pone.0002643. PLoS One. 2008. PMID: 18612465 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

