MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B
- PMID: 17890317
- PMCID: PMC2000384
- DOI: 10.1073/pnas.0707628104
MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B
Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate expression of many genes. Recent studies suggest roles of miRNAs in carcinogenesis. We and others have shown that expression profiles of miRNAs are different in lung cancer vs. normal lung, although the significance of this aberrant expression is poorly understood. Among the reported down-regulated miRNAs in lung cancer, the miRNA (miR)-29 family (29a, 29b, and 29c) has intriguing complementarities to the 3'-UTRs of DNA methyltransferase (DNMT)3A and -3B (de novo methyltransferases), two key enzymes involved in DNA methylation, that are frequently up-regulated in lung cancer and associated with poor prognosis. We investigated whether miR-29s could target DNMT3A and -B and whether restoration of miR-29s could normalize aberrant patterns of methylation in non-small-cell lung cancer. Here we show that expression of miR-29s is inversely correlated to DNMT3A and -3B in lung cancer tissues, and that miR-29s directly target both DNMT3A and -3B. The enforced expression of miR-29s in lung cancer cell lines restores normal patterns of DNA methylation, induces reexpression of methylation-silenced tumor suppressor genes, such as FHIT and WWOX, and inhibits tumorigenicity in vitro and in vivo. These findings support a role of miR-29s in epigenetic normalization of NSCLC, providing a rationale for the development of miRNA-based strategies for the treatment of lung cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



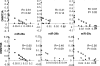

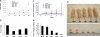
Similar articles
-
A regulatory circuitry comprising TP53, miR-29 family, and SETDB1 in non-small cell lung cancer.Biosci Rep. 2018 Sep 13;38(5):BSR20180678. doi: 10.1042/BSR20180678. Print 2018 Oct 31. Biosci Rep. 2018. PMID: 30054425 Free PMC article.
-
Suppression of Wnt signaling by the miR-29 family is mediated by demethylation of WIF-1 in non-small-cell lung cancer.Biochem Biophys Res Commun. 2013 Sep 6;438(4):673-9. doi: 10.1016/j.bbrc.2013.07.123. Epub 2013 Aug 9. Biochem Biophys Res Commun. 2013. PMID: 23939044
-
Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1.Hepatology. 2010 Jul;52(1):60-70. doi: 10.1002/hep.23660. Hepatology. 2010. PMID: 20578129
-
Regulation of expression of microRNAs by DNA methylation in lung cancer.Biomarkers. 2016 Nov;21(7):589-99. doi: 10.3109/1354750X.2016.1171906. Epub 2016 Apr 27. Biomarkers. 2016. PMID: 27122255 Review.
-
MicroRNA Methylation in Colorectal Cancer.Adv Exp Med Biol. 2016;937:109-22. doi: 10.1007/978-3-319-42059-2_6. Adv Exp Med Biol. 2016. PMID: 27573897 Review.
Cited by
-
Association of aberrant DNA methylation in Apc(min/+) mice with the epithelial-mesenchymal transition and Wnt/β-catenin pathways: genome-wide analysis using MeDIP-seq.Cell Biosci. 2015 May 27;5:24. doi: 10.1186/s13578-015-0013-2. eCollection 2015. Cell Biosci. 2015. PMID: 26101583 Free PMC article.
-
Mechanistic Roles of Noncoding RNAs in Lung Cancer Biology and Their Clinical Implications.Genet Res Int. 2012;2012:737416. doi: 10.1155/2012/737416. Epub 2012 Jul 18. Genet Res Int. 2012. PMID: 22852089 Free PMC article.
-
MicroRNAs and Glucocorticoid-Induced Apoptosis in Lymphoid Malignancies.ISRN Hematol. 2013;2013:348212. doi: 10.1155/2013/348212. Epub 2013 Jan 29. ISRN Hematol. 2013. PMID: 23431463 Free PMC article.
-
MicroRNA as Biomarkers and Diagnostics.J Cell Physiol. 2016 Jan;231(1):25-30. doi: 10.1002/jcp.25056. J Cell Physiol. 2016. PMID: 26031493 Free PMC article. Review.
-
Associations Between Epigenetic Age Acceleration and microRNA Expression Among U.S. Firefighters.Epigenet Insights. 2023 Nov 8;16:25168657231206301. doi: 10.1177/25168657231206301. eCollection 2023. Epigenet Insights. 2023. PMID: 37953967 Free PMC article.
References
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. CA Cancer J Clin. 2007;57:43–66. - PubMed
-
- Ulivi P, Zoli W, Calistri D, Fabbri F, Tesei A, Rosetti M, Mengozzi M, Amadori D. J Cell Physiol. 2006;206:611–615. - PubMed
-
- Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, Palazzo J, McCue PA, Baffa R, Huebner K. Oncogene. 2005;24:1625–1633. - PubMed
-
- Suzuki M, Sunaga N, Shames DS, Toyooka S, Gazdar AF, Minna JD. Cancer Res. 2004;64:3137–3143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

