PocketPicker: analysis of ligand binding-sites with shape descriptors
- PMID: 17880740
- PMCID: PMC1994066
- DOI: 10.1186/1752-153X-1-7
PocketPicker: analysis of ligand binding-sites with shape descriptors
Abstract
Background: Identification and evaluation of surface binding-pockets and occluded cavities are initial steps in protein structure-based drug design. Characterizing the active site's shape as well as the distribution of surrounding residues plays an important role for a variety of applications such as automated ligand docking or in situ modeling. Comparing the shape similarity of binding site geometries of related proteins provides further insights into the mechanisms of ligand binding.
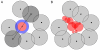
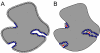
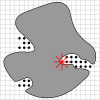
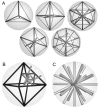
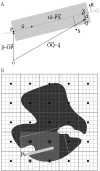
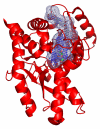
Results: We present PocketPicker, an automated grid-based technique for the prediction of protein binding pockets that specifies the shape of a potential binding-site with regard to its buriedness. The method was applied to a representative set of protein-ligand complexes and their corresponding apo-protein structures to evaluate the quality of binding-site predictions. The performance of the pocket detection routine was compared to results achieved with the existing methods CAST, LIGSITE, LIGSITE(cs), PASS and SURFNET. Success rates PocketPicker were comparable to those of LIGSITE(cs) and outperformed the other tools. We introduce a descriptor that translates the arrangement of grid points delineating a detected binding-site into a correlation vector. We show that this shape descriptor is suited for comparative analyses of similar binding-site geometry by examining induced-fit phenomena in aldose reductase. This new method uses information derived from calculations of the buriedness of potential binding-sites.
Conclusion: The pocket prediction routine of PocketPicker is a useful tool for identification of potential protein binding-pockets. It produces a convenient representation of binding-site shapes including an intuitive description of their accessibility. The shape-descriptor for automated classification of binding-site geometries can be used as an additional tool complementing elaborate manual inspections.
Figures
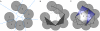

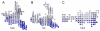



Similar articles
-
Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design.Protein Sci. 1998 Sep;7(9):1884-97. doi: 10.1002/pro.5560070905. Protein Sci. 1998. PMID: 9761470 Free PMC article.
-
Relating the shape of protein binding sites to binding affinity profiles: is there an association?BMC Struct Biol. 2010 Oct 5;10:32. doi: 10.1186/1472-6807-10-32. BMC Struct Biol. 2010. PMID: 20923553 Free PMC article.
-
Form follows function: shape analysis of protein cavities for receptor-based drug design.Proteomics. 2009 Jan;9(2):451-9. doi: 10.1002/pmic.200800092. Proteomics. 2009. PMID: 19142949
-
Protein Binding Pocket Dynamics.Acc Chem Res. 2016 May 17;49(5):809-15. doi: 10.1021/acs.accounts.5b00516. Epub 2016 Apr 25. Acc Chem Res. 2016. PMID: 27110726 Review.
-
Methods for the prediction of protein-ligand binding sites for structure-based drug design and virtual ligand screening.Curr Protein Pept Sci. 2006 Oct;7(5):395-406. doi: 10.2174/138920306778559386. Curr Protein Pept Sci. 2006. PMID: 17073692 Review.
Cited by
-
GPCR-BSD: a database of binding sites of human G-protein coupled receptors under diverse states.BMC Bioinformatics. 2024 Nov 4;25(1):343. doi: 10.1186/s12859-024-05962-9. BMC Bioinformatics. 2024. PMID: 39497074 Free PMC article.
-
Druggable protein interaction sites are more predisposed to surface pocket formation than the rest of the protein surface.PLoS Comput Biol. 2013;9(3):e1002951. doi: 10.1371/journal.pcbi.1002951. Epub 2013 Mar 7. PLoS Comput Biol. 2013. PMID: 23505360 Free PMC article.
-
Protein pocket detection via convex hull surface evolution and associated Reeb graph.Bioinformatics. 2018 Sep 1;34(17):i830-i837. doi: 10.1093/bioinformatics/bty598. Bioinformatics. 2018. PMID: 30423105 Free PMC article.
-
SmoPSI: Analysis and Prediction of Small Molecule Binding Sites Based on Protein Sequence Information.Comput Math Methods Med. 2019 Nov 13;2019:1926156. doi: 10.1155/2019/1926156. eCollection 2019. Comput Math Methods Med. 2019. PMID: 31814842 Free PMC article.
-
Identification of recurring protein structure microenvironments and discovery of novel functional sites around CYS residues.BMC Struct Biol. 2010 Feb 2;10:4. doi: 10.1186/1472-6807-10-4. BMC Struct Biol. 2010. PMID: 20122268 Free PMC article.
References
-
- Gane PJ, Dean PM. Recent advances in structure-based rational drug design. Curr Opin Struct Biol. 2000;10:401–404. - PubMed
-
- Klebe G. Recent developments in structure-based rational design. J Mol Med. 2000;78:269–281. - PubMed
-
- Sotriffer CA, Klebe G. Identification and mapping of small-molecule binding sites in proteins: computational tools for structure-based drug design. Farmaco. 2002;57:243–251. - PubMed
-
- Campbell SJ, Gold ND, Jackson RM, Westhead DR. Ligand binding: functional site location, similarity and docking. Curr Opin Struct Biol. 2003;13:389–395. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

