Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila
- PMID: 17803358
- PMCID: PMC1964775
- DOI: 10.1371/journal.pbio.0050238
Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila
Abstract
The activation of several transcription factors is required for the elimination of infectious pathogens via the innate immune response. The transcription factors NF-kappaB, AP-1, and STAT play major roles in the synthesis of immune effector molecules during innate immune responses. However, the fact that these immune responses can have cytotoxic effects requires their tight regulation to achieve restricted and transient activation, and mis-regulation of the damping process has pathological consequences. Here we show that AP-1 and STAT are themselves the major inhibitors responsible for damping NF-kappaB-mediated transcriptional activation during the innate immune response in Drosophila. As the levels of dAP-1 and Stat92E increase due to continuous immune signaling, they play a repressive role by forming a repressosome complex with the Drosophila HMG protein, Dsp1. The dAP-1-, Stat92E-, and Dsp1-containing complexes replace Relish at the promoters of diverse immune effector genes by binding to evolutionarily conserved cis-elements, and they recruit histone deacetylase to inhibit transcription. Reduction by mutation of dAP-1, Stat92E, or Dsp1 results in hyperactivation of Relish target genes and reduces the viability of bacterially infected flies despite more efficient pathogen clearance. These defects are rescued by reducing the Relish copy number, thus confirming that mis-regulation of Relish, not inadequate activation of dAP-1, Stat92E, or Dsp1 target genes, is responsible for the reduced survival of the mutants. We conclude that an inhibitory effect of AP-1 and STAT on NF-kappaB is required for properly balanced immune responses and appears to be evolutionarily conserved.
Conflict of interest statement
Figures

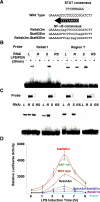
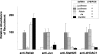
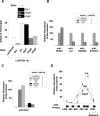
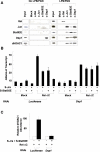
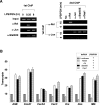
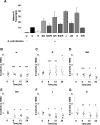
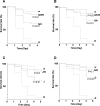
Comment in
-
How and why does a fly turn its immune system off?PLoS Biol. 2007 Sep;5(9):e247. doi: 10.1371/journal.pbio.0050247. PLoS Biol. 2007. PMID: 17880266 Free PMC article.
Similar articles
-
Akirin specifies NF-κB selectivity of Drosophila innate immune response via chromatin remodeling.EMBO J. 2014 Oct 16;33(20):2349-62. doi: 10.15252/embj.201488456. Epub 2014 Sep 1. EMBO J. 2014. PMID: 25180232 Free PMC article.
-
Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP1 and NF-kappaB signaling modules.Nat Immunol. 2005 Feb;6(2):211-8. doi: 10.1038/ni1159. Epub 2005 Jan 9. Nat Immunol. 2005. PMID: 15640802
-
Osa-Containing Brahma Complex Regulates Innate Immunity and the Expression of Metabolic Genes in Drosophila.J Immunol. 2020 Apr 15;204(8):2143-2155. doi: 10.4049/jimmunol.1900571. Epub 2020 Mar 20. J Immunol. 2020. PMID: 32198143
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
-
Antagonistic crosstalk between NF-κB and SIRT1 in the regulation of inflammation and metabolic disorders.Cell Signal. 2013 Oct;25(10):1939-48. doi: 10.1016/j.cellsig.2013.06.007. Epub 2013 Jun 11. Cell Signal. 2013. PMID: 23770291 Review.
Cited by
-
Ebi, a Drosophila homologue of TBL1, regulates the balance between cellular defense responses and neuronal survival.Am J Neurodegener Dis. 2016 Mar 1;5(1):62-8. eCollection 2016. Am J Neurodegener Dis. 2016. PMID: 27073743 Free PMC article.
-
Dynamic Regulation of NF-κB Response in Innate Immunity: The Case of the IMD Pathway in Drosophila.Biomedicines. 2022 Sep 16;10(9):2304. doi: 10.3390/biomedicines10092304. Biomedicines. 2022. PMID: 36140409 Free PMC article. Review.
-
Rab35 mediates transport of Cdc42 and Rac1 to the plasma membrane during phagocytosis.Mol Cell Biol. 2010 Mar;30(6):1421-33. doi: 10.1128/MCB.01463-09. Epub 2010 Jan 11. Mol Cell Biol. 2010. PMID: 20065041 Free PMC article.
-
SUMOylation of Jun fine-tunes the Drosophila gut immune response.PLoS Pathog. 2022 Mar 7;18(3):e1010356. doi: 10.1371/journal.ppat.1010356. eCollection 2022 Mar. PLoS Pathog. 2022. PMID: 35255103 Free PMC article.
-
Molecular architecture of the fruit fly's airway epithelial immune system.BMC Genomics. 2008 Sep 29;9:446. doi: 10.1186/1471-2164-9-446. BMC Genomics. 2008. PMID: 18823557 Free PMC article.
References
-
- Janeway CA, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. - PubMed
-
- Kimbrell DA, Beutler B. The evolution and genetics of innate immunity. Nat Rev Genet. 2001;2:256–267. - PubMed
-
- Kim T, Kim Y-J. Overview of innate immunity in Drosophila . J Biochem Mol Biol. 2005;38:121–127. - PubMed
-
- Silverman N, Maniatis T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15:2321–2342. - PubMed
-
- Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, et al. Relish, a central factor in the control of humoral, but not cellular immunity in Drosophila . Mol Cell. 1999;4:827–837. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

