SUMO-targeted ubiquitin ligases in genome stability
- PMID: 17762865
- PMCID: PMC2230673
- DOI: 10.1038/sj.emboj.7601838
SUMO-targeted ubiquitin ligases in genome stability
Abstract
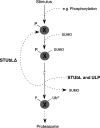
We identify the SUMO-Targeted Ubiquitin Ligase (STUbL) family of proteins and propose that STUbLs selectively ubiquitinate sumoylated proteins and proteins that contain SUMO-like domains (SLDs). STUbL recruitment to sumoylated/SLD proteins is mediated by tandem SUMO interaction motifs (SIMs) within the STUbLs N-terminus. STUbL-mediated ubiquitination maintains sumoylation pathway homeostasis by promoting target protein desumoylation and/or degradation. Thus, STUbLs establish a novel mode of communication between the sumoylation and ubiquitination pathways. STUbLs are evolutionarily conserved and include: Schizosaccharomyces pombe Slx8-Rfp (founding member), Homo sapiens RNF4, Dictyostelium discoideum MIP1 and Saccharomyces cerevisiae Slx5-Slx8. Cells lacking Slx8-Rfp accumulate sumoylated proteins, display genomic instability, and are hypersensitive to genotoxic stress. These phenotypes are suppressed by deletion of the major SUMO ligase Pli1, demonstrating the specificity of STUbLs as regulators of sumoylated proteins. Notably, human RNF4 expression restores SUMO pathway homeostasis in fission yeast lacking Slx8-Rfp, underscoring the evolutionary functional conservation of STUbLs. The DNA repair factor Rad60 and its human homolog NIP45, which contain SLDs, are candidate STUbL targets. Consistently, Rad60 and Slx8-Rfp mutants have similar DNA repair defects.
Figures



Similar articles
-
Functional Crosstalk between the PP2A and SUMO Pathways Revealed by Analysis of STUbL Suppressor, razor 1-1.PLoS Genet. 2016 Jul 11;12(7):e1006165. doi: 10.1371/journal.pgen.1006165. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27398807 Free PMC article.
-
Concerted action of the ubiquitin-fusion degradation protein 1 (Ufd1) and Sumo-targeted ubiquitin ligases (STUbLs) in the DNA-damage response.PLoS One. 2013 Nov 12;8(11):e80442. doi: 10.1371/journal.pone.0080442. eCollection 2013. PLoS One. 2013. PMID: 24265825 Free PMC article.
-
Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins.EMBO J. 2007 Sep 19;26(18):4102-12. doi: 10.1038/sj.emboj.7601839. Epub 2007 Aug 30. EMBO J. 2007. PMID: 17762864 Free PMC article.
-
A SIM-ultaneous role for SUMO and ubiquitin.Trends Biochem Sci. 2008 May;33(5):201-8. doi: 10.1016/j.tibs.2008.02.001. Epub 2008 Apr 9. Trends Biochem Sci. 2008. PMID: 18403209 Review.
-
SUMO-targeted ubiquitin ligases.Biochim Biophys Acta. 2014 Jan;1843(1):75-85. doi: 10.1016/j.bbamcr.2013.08.022. Epub 2013 Sep 7. Biochim Biophys Acta. 2014. PMID: 24018209 Review.
Cited by
-
RNF4 is required for DNA double-strand break repair in vivo.Cell Death Differ. 2013 Mar;20(3):490-502. doi: 10.1038/cdd.2012.145. Epub 2012 Nov 30. Cell Death Differ. 2013. PMID: 23197296 Free PMC article.
-
Ubiquitin-specific Protease 11 (USP11) Deubiquitinates Hybrid Small Ubiquitin-like Modifier (SUMO)-Ubiquitin Chains to Counteract RING Finger Protein 4 (RNF4).J Biol Chem. 2015 Jun 19;290(25):15526-15537. doi: 10.1074/jbc.M114.618132. Epub 2015 May 12. J Biol Chem. 2015. PMID: 25969536 Free PMC article.
-
PARP-1 transcriptional activity is regulated by sumoylation upon heat shock.EMBO J. 2009 Nov 18;28(22):3534-48. doi: 10.1038/emboj.2009.279. Epub 2009 Sep 24. EMBO J. 2009. PMID: 19779455 Free PMC article.
-
Cardiac function and disease: emerging role of small ubiquitin-related modifier.Wiley Interdiscip Rev Syst Biol Med. 2011 Jul-Aug;3(4):446-57. doi: 10.1002/wsbm.130. Epub 2010 Dec 31. Wiley Interdiscip Rev Syst Biol Med. 2011. PMID: 21197655 Free PMC article. Review.
-
RNF4 and USP7 cooperate in ubiquitin-regulated steps of DNA replication.Open Biol. 2023 Aug;13(8):230068. doi: 10.1098/rsob.230068. Epub 2023 Aug 23. Open Biol. 2023. PMID: 37607592 Free PMC article.
References
-
- Abdel-Hafiz H, Takimoto GS, Tung L, Horwitz KB (2002) The inhibitory function in human progesterone receptor N termini binds SUMO-1 protein to regulate autoinhibition and transrepression. J Biol Chem 277: 33950–33956 - PubMed
-
- al-Khodairy F, Enoch T, Hagan IM, Carr AM (1995) The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J Cell Sci 108 (Part 2): 475–486 - PubMed
-
- Boddy MN, Furnari B, Mondesert O, Russell P (1998) Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280: 909–912 - PubMed
-
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR III, Russell P (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

