Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus
- PMID: 17761676
- PMCID: PMC2756044
- DOI: 10.1074/jbc.M704870200
Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus
Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV) is a novel coronavirus that causes a highly contagious respiratory disease, SARS, with significant mortality. Although factors contributing to the highly pathogenic nature of SARS-CoV remain poorly understood, it has been reported that SARS-CoV infection does not induce type I interferons (IFNs) in cell culture. However, it is uncertain whether SARS-CoV evades host detection or has evolved mechanisms to counteract innate host defenses. We show here that infection of SARS-CoV triggers a weak IFN response in cultured human lung/bronchial epithelial cells without inducing the phosphorylation of IFN-regulatory factor 3 (IRF-3), a latent cellular transcription factor that is pivotal for type I IFN synthesis. Furthermore, SARS-CoV infection blocked the induction of IFN antiviral activity and the up-regulation of protein expression of a subset of IFN-stimulated genes triggered by double-stranded RNA or an unrelated paramyxovirus. In searching for a SARS-CoV protein capable of counteracting innate immunity, we identified the papain-like protease (PLpro) domain as a potent IFN antagonist. The inhibition of the IFN response does not require the protease activity of PLpro. Rather, PLpro interacts with IRF-3 and inhibits the phosphorylation and nuclear translocation of IRF-3, thereby disrupting the activation of type I IFN responses through either Toll-like receptor 3 or retinoic acid-inducible gene I/melanoma differentiation-associated gene 5 pathways. Our data suggest that regulation of IRF-3-dependent innate antiviral defenses by PLpro may contribute to the establishment of SARS-CoV infection.
Figures

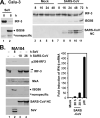
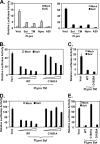
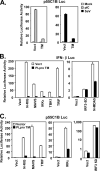
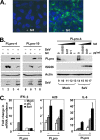

Similar articles
-
Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling.PLoS One. 2012;7(2):e30802. doi: 10.1371/journal.pone.0030802. Epub 2012 Feb 1. PLoS One. 2012. PMID: 22312431 Free PMC article.
-
SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex.Protein Cell. 2014 May;5(5):369-81. doi: 10.1007/s13238-014-0026-3. Epub 2014 Mar 14. Protein Cell. 2014. PMID: 24622840 Free PMC article.
-
Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection.PLoS One. 2010 Jan 15;5(1):e8729. doi: 10.1371/journal.pone.0008729. PLoS One. 2010. PMID: 20090954 Free PMC article.
-
The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds.Antiviral Res. 2015 Mar;115:21-38. doi: 10.1016/j.antiviral.2014.12.015. Epub 2014 Dec 29. Antiviral Res. 2015. PMID: 25554382 Free PMC article. Review.
-
Coronavirus virulence genes with main focus on SARS-CoV envelope gene.Virus Res. 2014 Dec 19;194:124-37. doi: 10.1016/j.virusres.2014.07.024. Epub 2014 Aug 2. Virus Res. 2014. PMID: 25093995 Free PMC article. Review.
Cited by
-
Viral Factors in Modulation of Host Immune Response: A Route to Novel Antiviral Agents and New Therapeutic Approaches.Int J Mol Sci. 2024 Aug 29;25(17):9408. doi: 10.3390/ijms25179408. Int J Mol Sci. 2024. PMID: 39273355 Free PMC article. Review.
-
Profiling of coronaviral Mpro and enteroviral 3Cpro specificity provides a framework for the development of broad-spectrum antiviral compounds.Protein Sci. 2024 Sep;33(9):e5139. doi: 10.1002/pro.5139. Protein Sci. 2024. PMID: 39150063
-
Viral deubiquitinating proteases and the promising strategies of their inhibition.Virus Res. 2024 Jun;344:199368. doi: 10.1016/j.virusres.2024.199368. Epub 2024 Apr 11. Virus Res. 2024. PMID: 38588924 Free PMC article. Review.
-
USP2 inhibition prevents infection with ACE2-dependent coronaviruses in vitro and is protective against SARS-CoV-2 in mice.Sci Transl Med. 2023 Dec 6;15(725):eadh7668. doi: 10.1126/scitranslmed.adh7668. Epub 2023 Dec 6. Sci Transl Med. 2023. PMID: 38055802
-
Targeting SARS-CoV-2 nonstructural protein 3: Function, structure, inhibition, and perspective in drug discovery.Drug Discov Today. 2024 Jan;29(1):103832. doi: 10.1016/j.drudis.2023.103832. Epub 2023 Nov 15. Drug Discov Today. 2024. PMID: 37977285 Review.
References
-
- Sen G.C. Annu. Rev. Microbiol. 2001;55:255–281. - PubMed
-
- Goutagny N., Kim M., Fitzgerald K.A. Nat. Immunol. 2006;7:555–557. - PubMed
-
- Stetson D.B., Kim R. Immunity. 2006;25:373–381. - PubMed
-
- Garcia-Sastre A., Kim C.A. Science. 2006;312:879–882. - PubMed
-
- Kawai T., Kim S. Nat. Immunol. 2006;7:131–137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DA018054/DA/NIDA NIH HHS/United States
- P01 AI060915-030001/AI/NIAID NIH HHS/United States
- R21 DA018054-01/DA/NIDA NIH HHS/United States
- AI45798/AI/NIAID NIH HHS/United States
- N01 AI030039-009/AI/NIAID NIH HHS/United States
- R01 AI045798/AI/NIAID NIH HHS/United States
- P01 AI060915/AI/NIAID NIH HHS/United States
- AI057156/AI/NIAID NIH HHS/United States
- U54 AI057156-05S10038/AI/NIAID NIH HHS/United States
- R01 AI069285/AI/NIAID NIH HHS/United States
- AI060915/AI/NIAID NIH HHS/United States
- R01 AI069285-01A2/AI/NIAID NIH HHS/United States
- U54 AI057156/AI/NIAID NIH HHS/United States
- AI30039/AI/NIAID NIH HHS/United States
- AI069285/AI/NIAID NIH HHS/United States
- R21 DA018054/DA/NIDA NIH HHS/United States
- R01 AI045798-01A2/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

