Cytotoxic ribonucleases: the dichotomy of Coulombic forces
- PMID: 17705507
- PMCID: PMC2864629
- DOI: 10.1021/bi700857u
Cytotoxic ribonucleases: the dichotomy of Coulombic forces
Abstract
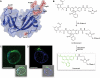
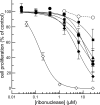
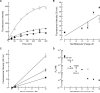
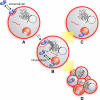
Cells tightly regulate their contents. Still, nonspecific Coulombic interactions between cationic molecules and anionic membrane components can lead to adventitious endocytosis. Here, we characterize this process in a natural system. To do so, we create variants of human pancreatic ribonuclease (RNase 1) that differ in net molecular charge. By conjugating a small-molecule latent fluorophore to these variants and using flow cytometry, we are able to determine the kinetic mechanism for RNase 1 internalization into live human cells. We find that internalization increases with solution concentration and is not saturable. Internalization also increases with time to a steady-state level, which varies linearly with molecular charge. In contrast, the rate constant for internalization (t1/2 = 2 h) is independent of charge. We conclude that internalization involves an extracellular equilibrium complex between the cationic proteins and abundant anionic cell-surface molecules, followed by rate-limiting internalization. The enhanced internalization of more cationic variants of RNase 1 is, however, countered by their increased affinity for the cytosolic ribonuclease inhibitor protein, which is anionic. Thus, Coulombic forces mediate extracellular and intracellular equilibria in a dichotomous manner that both endangers cells and defends them from the potentially lethal enzymatic activity of ribonucleases.
Figures

Similar articles
-
Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein.J Mol Biol. 2007 Apr 27;368(2):434-49. doi: 10.1016/j.jmb.2007.02.005. Epub 2007 Feb 9. J Mol Biol. 2007. PMID: 17350650 Free PMC article.
-
Endocytotic internalization as a crucial factor for the cytotoxicity of ribonucleases.J Biol Chem. 2007 Sep 21;282(38):27640-6. doi: 10.1074/jbc.M702240200. Epub 2007 Jul 17. J Biol Chem. 2007. PMID: 17635931
-
Tandemization endows bovine pancreatic ribonuclease with cytotoxic activity.J Mol Biol. 2006 May 19;358(5):1305-13. doi: 10.1016/j.jmb.2006.03.007. Epub 2006 Mar 21. J Mol Biol. 2006. PMID: 16580680
-
Design of cytotoxic ribonucleases by cationization to enhance intracellular protein delivery.Curr Pharm Biotechnol. 2008 Jun;9(3):180-4. doi: 10.2174/138920108784567326. Curr Pharm Biotechnol. 2008. PMID: 18673283 Review.
-
Structural and functional relationships of natural and artificial dimeric bovine ribonucleases: new scaffolds for potential antitumor drugs.FEBS Lett. 2013 Nov 15;587(22):3601-8. doi: 10.1016/j.febslet.2013.09.038. Epub 2013 Oct 7. FEBS Lett. 2013. PMID: 24113657 Review.
Cited by
-
A highly sensitive fluorogenic probe for cytochrome P450 activity in live cells.Bioorg Med Chem Lett. 2008 Nov 15;18(22):5864-6. doi: 10.1016/j.bmcl.2008.06.015. Epub 2008 Jun 10. Bioorg Med Chem Lett. 2008. PMID: 18595692 Free PMC article.
-
Mechanism of ribonuclease A endocytosis: analogies to cell-penetrating peptides.Biochemistry. 2011 Oct 4;50(39):8374-82. doi: 10.1021/bi2009079. Epub 2011 Sep 7. Biochemistry. 2011. PMID: 21827164 Free PMC article.
-
Antitumor activity of ribonuclease multimers created by site-specific covalent tethering.Bioconjug Chem. 2010 Sep 15;21(9):1691-702. doi: 10.1021/bc100292x. Bioconjug Chem. 2010. PMID: 20704261 Free PMC article.
-
Cellular uptake of ribonuclease A relies on anionic glycans.Biochemistry. 2010 Dec 21;49(50):10666-73. doi: 10.1021/bi1013485. Epub 2010 Nov 23. Biochemistry. 2010. PMID: 21062061 Free PMC article.
-
Onconase and amphinase, the antitumor ribonucleases from Rana pipiens oocytes.Curr Pharm Biotechnol. 2008 Jun;9(3):215-25. doi: 10.2174/138920108784567245. Curr Pharm Biotechnol. 2008. PMID: 18673287 Free PMC article. Review.
References
-
- Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. - PubMed
-
- Polo S, Di Fiore PP. Endocytosis conducts the cell signaling orchestra. Cell. 2006;124:897–900. - PubMed
-
- Murray D, Honig B. Electrostatic control of the membrane targeting of C2 domains. Mol. Cell. 2002;9:145–54. - PubMed
-
- Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005;34:119–51. - PubMed
-
- Mulgrew-Nesbitt A, Diraviyam K, Wang J, Singh S, Murray P, Li Z, Rogers L, Mirkovic N, Murray D. The role of electrostatics in protein-membrane interactions. Biochim. Biophys. Acta. 2006;1761:812–826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

