Stimulation of in vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway
- PMID: 17669696
- PMCID: PMC2100399
- DOI: 10.1016/j.dnarep.2007.06.004
Stimulation of in vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway
Abstract
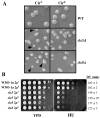
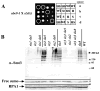
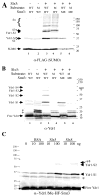
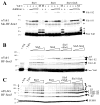
The yeast genes SLX5 and SLX8 were identified based on their requirement for viability in the absence of the Sgs1 DNA helicase. Loss of these genes results in genome instability, nibbled colonies, and other phenotypes associated with defects in sumoylation. The Slx5 and Slx8 proteins form a stable complex and each subunit contains a single RING-finger domain at its C-terminus. To determine the physiological function of the Slx5-8 complex, we explored its interaction with the SUMO pathway. Curing 2micro circle from the mutants, suppressed their nibbled colony phenotype and partially improved their growth rate, but did not affect their sensitivity to hydroxyurea. The increase in sumoylation observed in slx5Delta and slx8Delta mutants was found to be dependent on the Siz1 SUMO ligase. Physical interactions between the Slx5-8 complex and both Ubc9 and Smt3 were identified and characterized. Using in vitro reactions, we show that Slx5, Slx8, or the Slx5-8 complex stimulates the formation of SUMO chains and the sumoylation of a test substrate. Interestingly, a functional RING-finger domain is not required for this stimulation in vitro. These biochemical data demonstrate for the first time that the Slx5 and Slx8 complex is capable of interacting directly with the SUMO pathway.
Figures



Similar articles
-
Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates.J Biol Chem. 2008 Jul 18;283(29):19912-21. doi: 10.1074/jbc.M802690200. Epub 2008 May 22. J Biol Chem. 2008. PMID: 18499666 Free PMC article.
-
Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity.Nucleic Acids Res. 2006;34(19):5541-51. doi: 10.1093/nar/gkl685. Epub 2006 Oct 4. Nucleic Acids Res. 2006. PMID: 17020915 Free PMC article.
-
A Lysine Desert Protects a Novel Domain in the Slx5-Slx8 SUMO Targeted Ub Ligase To Maintain Sumoylation Levels in Saccharomyces cerevisiae.Genetics. 2017 Aug;206(4):1807-1821. doi: 10.1534/genetics.117.202697. Epub 2017 May 26. Genetics. 2017. PMID: 28550017 Free PMC article.
-
SUMO-targeted ubiquitin ligases.Biochim Biophys Acta. 2014 Jan;1843(1):75-85. doi: 10.1016/j.bbamcr.2013.08.022. Epub 2013 Sep 7. Biochim Biophys Acta. 2014. PMID: 24018209 Review.
-
Nuclear organization in genome stability: SUMO connections.Cell Res. 2011 Mar;21(3):474-85. doi: 10.1038/cr.2011.31. Epub 2011 Feb 15. Cell Res. 2011. PMID: 21321608 Free PMC article. Review.
Cited by
-
Genome stability roles of SUMO-targeted ubiquitin ligases.DNA Repair (Amst). 2009 Apr 5;8(4):517-24. doi: 10.1016/j.dnarep.2009.01.010. Epub 2009 Feb 23. DNA Repair (Amst). 2009. PMID: 19233742 Free PMC article. Review.
-
Slx5 promotes transcriptional silencing and is required for robust growth in the absence of Sir2.Mol Cell Biol. 2008 Feb;28(4):1361-72. doi: 10.1128/MCB.01291-07. Epub 2007 Dec 17. Mol Cell Biol. 2008. PMID: 18086879 Free PMC article.
-
A Flp-SUMO hybrid recombinase reveals multi-layered copy number control of a selfish DNA element through post-translational modification.PLoS Genet. 2019 Jun 26;15(6):e1008193. doi: 10.1371/journal.pgen.1008193. eCollection 2019 Jun. PLoS Genet. 2019. PMID: 31242181 Free PMC article.
-
Wss1 metalloprotease partners with Cdc48/Doa1 in processing genotoxic SUMO conjugates.Elife. 2015 Sep 8;4:e06763. doi: 10.7554/eLife.06763. Elife. 2015. PMID: 26349035 Free PMC article.
-
The yeast Slx5-Slx8 DNA integrity complex displays ubiquitin ligase activity.Cell Cycle. 2007 Nov 15;6(22):2800-9. doi: 10.4161/cc.6.22.4882. Epub 2007 Aug 13. Cell Cycle. 2007. PMID: 18032921 Free PMC article.
References
-
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. - PubMed
-
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. - PubMed
-
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. - PubMed
-
- Stade K, Vogel F, Schwienhorst I, Meusser B, Volkwein C, Nentwig B, Dohmen RJ, Sommer T. A lack of SUMO conjugation affects cNLS-dependent nuclear protein import in yeast. J Biol Chem. 2002;277:49554–49561. - PubMed
-
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol Cell. 1998;2:233–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

