Structural and functional analysis of RNA and TAP binding to SF2/ASF
- PMID: 17668007
- PMCID: PMC1978082
- DOI: 10.1038/sj.embor.7401031
Structural and functional analysis of RNA and TAP binding to SF2/ASF
Abstract
The serine/arginine-rich (SR) protein splicing factor 2/alternative splicing factor (SF2/ASF) has a role in splicing, stability, export and translation of messenger RNA. Here, we present the structure of the RNA recognition motif (RRM) 2 from SF2/ASF, which has an RRM fold with a considerably extended loop 5 region, containing a two-stranded beta-sheet. The loop 5 extension places the previously identified SR protein kinase 1 docking sequence largely within the RRM fold. We show that RRM2 binds to RNA in a new way, by using a tryptophan within a conserved SWQLKD motif that resides on helix alpha1, together with amino acids from strand beta2 and a histidine on loop 5. The linker connecting RRM1 and RRM2 contains arginine residues, which provide a binding site for the mRNA export factor TAP, and when TAP binds to this region it displaces RNA bound to RRM2.
Figures
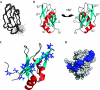


Similar articles
-
Regiospecific phosphorylation control of the SR protein ASF/SF2 by SRPK1.J Mol Biol. 2009 Jul 24;390(4):618-34. doi: 10.1016/j.jmb.2009.05.060. Epub 2009 May 27. J Mol Biol. 2009. PMID: 19477182 Free PMC article.
-
The second RNA-binding domain of the human splicing factor ASF/SF2 is the critical domain controlling adenovirus E1A alternative 5'-splice site selection.Biochem J. 2004 Jul 15;381(Pt 2):343-50. doi: 10.1042/BJ20040408. Biochem J. 2004. PMID: 15068396 Free PMC article.
-
Deletion of the N-terminus of SF2/ASF permits RS-domain-independent pre-mRNA splicing.PLoS One. 2007 Sep 5;2(9):e854. doi: 10.1371/journal.pone.0000854. PLoS One. 2007. PMID: 17786225 Free PMC article.
-
Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2.Mol Cell. 2005 Oct 7;20(1):77-89. doi: 10.1016/j.molcel.2005.08.025. Mol Cell. 2005. PMID: 16209947
-
A serine/arginine-rich domain in the human U1 70k protein is necessary and sufficient for ASF/SF2 binding.J Biol Chem. 1998 Aug 7;273(32):20629-35. doi: 10.1074/jbc.273.32.20629. J Biol Chem. 1998. PMID: 9685421
Cited by
-
Conserved proline-directed phosphorylation regulates SR protein conformation and splicing function.Biochem J. 2015 Mar 1;466(2):311-22. doi: 10.1042/BJ20141373. Biochem J. 2015. PMID: 25529026 Free PMC article.
-
The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation.Nat Struct Mol Biol. 2012 Jan 15;19(2):220-8. doi: 10.1038/nsmb.2207. Nat Struct Mol Biol. 2012. PMID: 22245967 Free PMC article.
-
Nuclear mRNA maturation and mRNA export control: from trypanosomes to opisthokonts.Parasitology. 2021 Sep;148(10):1196-1218. doi: 10.1017/S0031182021000068. Epub 2021 Jan 19. Parasitology. 2021. PMID: 33461637 Free PMC article. Review.
-
Structural basis for regulation of RNA-binding proteins by phosphorylation.ACS Chem Biol. 2015 Mar 20;10(3):652-66. doi: 10.1021/cb500860x. Epub 2015 Jan 14. ACS Chem Biol. 2015. PMID: 25535763 Free PMC article. Review.
-
Protein kinase a-dependent phosphorylation of serine 119 in the proto-oncogenic serine/arginine-rich splicing factor 1 modulates its activity as a splicing enhancer protein.Genes Cancer. 2011 Aug;2(8):841-51. doi: 10.1177/1947601911430226. Genes Cancer. 2011. PMID: 22393468 Free PMC article.
References
-
- Allain FH, Gilbert DE, Bouvet P, Feigon J (2000) Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. J Mol Biol 303: 227–241 - PubMed
-
- Cartegni L, Krainer AR (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30: 377–384 - PubMed
-
- Fribourg S, Gatfield D, Izaurralde E, Conti E (2003) A novel mode of RBD-protein recognition in the Y14–Mago complex. Nat Struct Biol 10: 433–439 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

