Unique genes in giant viruses: regular substitution pattern and anomalously short size
- PMID: 17652424
- PMCID: PMC1950904
- DOI: 10.1101/gr.6358607
Unique genes in giant viruses: regular substitution pattern and anomalously short size
Abstract
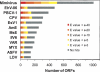
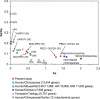
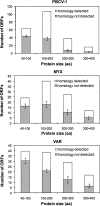
Large DNA viruses, including giant mimivirus with a 1.2-Mb genome, exhibit numerous orphan genes possessing no database homologs or genes with homologs solely in close members of the same viral family. Due to their solitary nature, the functions and evolutionary origins of those genes remain obscure. We examined sequence features and evolutionary rates of viral family-specific genes in three nucleo-cytoplasmic large DNA virus (NCLDV) lineages. First, we showed that the proportion of family-specific genes does not correlate with sequence divergence rate. Second, position-dependent nucleotide statistics were similar between family-specific genes and the remaining genes in the genome. Third, we showed that the synonymous-to-nonsynonymous substitution ratios in those viruses are at levels comparable to those estimated for vertebrate proteomes. Thus, the vast majority of family-specific genes do not exhibit an accelerated evolutionary rate, and are thus likely to specify functional polypeptides. On the other hand, these family-specific proteins exhibit several distinct properties: (1) they are shorter, (2) they include a larger fraction of predicted transmembrane proteins, and (3) they are enriched in low-complexity sequences. These results suggest that family-specific genes do not correspond to recent horizontal gene transfer. We propose that their characteristic features are the consequences of the specific evolutionary forces shaping the viral gene repertoires in the context of their parasitic lifestyles.
Figures



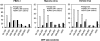
Similar articles
-
The 1.2-megabase genome sequence of Mimivirus.Science. 2004 Nov 19;306(5700):1344-50. doi: 10.1126/science.1101485. Epub 2004 Oct 14. Science. 2004. PMID: 15486256
-
Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review.
-
Structural and functional insights into Mimivirus ORFans.BMC Genomics. 2007 May 9;8:115. doi: 10.1186/1471-2164-8-115. BMC Genomics. 2007. PMID: 17490476 Free PMC article.
-
Convergent mechanisms of genome evolution of large and giant DNA viruses.Res Microbiol. 2008 Jun;159(5):325-31. doi: 10.1016/j.resmic.2008.04.012. Epub 2008 May 10. Res Microbiol. 2008. PMID: 18572389
-
Gene repertoire of amoeba-associated giant viruses.Intervirology. 2010;53(5):330-43. doi: 10.1159/000312918. Epub 2010 Jun 15. Intervirology. 2010. PMID: 20551685 Review.
Cited by
-
Genomics of Megavirus and the elusive fourth domain of Life.Commun Integr Biol. 2012 Jan 1;5(1):102-6. doi: 10.4161/cib.18624. Commun Integr Biol. 2012. PMID: 22482024 Free PMC article.
-
Horizontal gene transfer and nucleotide compositional anomaly in large DNA viruses.BMC Genomics. 2007 Dec 10;8:456. doi: 10.1186/1471-2164-8-456. BMC Genomics. 2007. PMID: 18070355 Free PMC article.
-
Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.Proteins. 2014 Feb;82 Suppl 2(0 2):26-42. doi: 10.1002/prot.24489. Proteins. 2014. PMID: 24318984 Free PMC article.
-
Two new subfamilies of DNA mismatch repair proteins (MutS) specifically abundant in the marine environment.ISME J. 2011 Jul;5(7):1143-51. doi: 10.1038/ismej.2010.210. Epub 2011 Jan 20. ISME J. 2011. PMID: 21248859 Free PMC article.
-
Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms.Front Cell Infect Microbiol. 2012 Aug 21;2:110. doi: 10.3389/fcimb.2012.00110. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919697 Free PMC article. Review.
References
-
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. - PMC - PubMed
-
- Angly F.E., Felts B., Breitbart M., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Felts B., Breitbart M., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Breitbart M., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Chan A.M., Haynes M., Kelley S., Liu H., Haynes M., Kelley S., Liu H., Kelley S., Liu H., Liu H., et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. - DOI - PMC - PubMed
-
- Benson S.D., Bamford J.K., Bamford D.H., Burnett R.M., Bamford J.K., Bamford D.H., Burnett R.M., Bamford D.H., Burnett R.M., Burnett R.M. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell. 2004;16:673–685. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
