Optimized design and assessment of whole genome tiling arrays
- PMID: 17646297
- PMCID: PMC5892713
- DOI: 10.1093/bioinformatics/btm200
Optimized design and assessment of whole genome tiling arrays
Abstract
Motivation: Recent advances in microarray technologies have made it feasible to interrogate whole genomes with tiling arrays and this technique is rapidly becoming one of the most important high-throughput functional genomics assays. For large mammalian genomes, analyzing oligonucleotide tiling array data is complicated by the presence of non-unique sequences on the array, which increases the overall noise in the data and may lead to false positive results due to cross-hybridization. The ability to create custom microarrays using maskless array synthesis has led us to consider ways to optimize array design characteristics for improving data quality and analysis. We have identified a number of design parameters to be optimized including uniqueness of the probe sequences within the whole genome, melting temperature and self-hybridization potential.
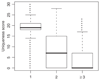
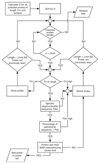
Results: We introduce the uniqueness score, U, a novel quality measure for oligonucleotide probes and present a method to quickly compute it. We show that U is equivalent to the number of shortest unique substrings in the probe and describe an efficient greedy algorithm to design mammalian whole genome tiling arrays using probes that maximize U. Using the mouse genome, we demonstrate how several optimizations influence the tiling array design characteristics. With a sensible set of parameters, our designs cover 78% of the mouse genome including many regions previously considered 'untilable' due to the presence of repetitive sequence. Finally, we compare our whole genome tiling array designs with commercially available designs.
Availability: Source code is available under an open source license from http://www.ebi.ac.uk/~graef/arraydesign/.
Conflict of interest statement
Figures


Similar articles
-
Custom design and analysis of high-density oligonucleotide bacterial tiling microarrays.PLoS One. 2009 Jun 17;4(6):e5943. doi: 10.1371/journal.pone.0005943. PLoS One. 2009. PMID: 19536279 Free PMC article.
-
Efficient algorithms for the computational design of optimal tiling arrays.IEEE/ACM Trans Comput Biol Bioinform. 2008 Oct-Dec;5(4):557-67. doi: 10.1109/TCBB.2008.50. IEEE/ACM Trans Comput Biol Bioinform. 2008. PMID: 18989043
-
Picky: oligo microarray design for large genomes.Bioinformatics. 2004 Nov 22;20(17):2893-902. doi: 10.1093/bioinformatics/bth347. Epub 2004 Jun 4. Bioinformatics. 2004. PMID: 15180932
-
In-depth query of large genomes using tiling arrays.Methods Mol Biol. 2007;377:163-74. doi: 10.1007/978-1-59745-390-5_10. Methods Mol Biol. 2007. PMID: 17634616 Review.
-
Applications of DNA tiling arrays to experimental genome annotation and regulatory pathway discovery.Chromosome Res. 2005;13(3):259-74. doi: 10.1007/s10577-005-2165-0. Chromosome Res. 2005. PMID: 15868420 Review.
Cited by
-
Ensembl 2009.Nucleic Acids Res. 2009 Jan;37(Database issue):D690-7. doi: 10.1093/nar/gkn828. Epub 2008 Nov 25. Nucleic Acids Res. 2009. PMID: 19033362 Free PMC article.
-
Sequence characteristics define trade-offs between on-target and genome-wide off-target hybridization of oligoprobes.PLoS One. 2018 Jun 21;13(6):e0199162. doi: 10.1371/journal.pone.0199162. eCollection 2018. PLoS One. 2018. PMID: 29928000 Free PMC article.
-
Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation.Mol Cell. 2010 May 28;38(4):603-13. doi: 10.1016/j.molcel.2010.03.016. Mol Cell. 2010. PMID: 20513434 Free PMC article.
-
Early life adversity alters normal sex-dependent developmental dynamics of DNA methylation.Dev Psychopathol. 2016 Nov;28(4pt2):1259-1272. doi: 10.1017/S0954579416000833. Epub 2016 Sep 30. Dev Psychopathol. 2016. PMID: 27687908 Free PMC article.
-
Custom design and analysis of high-density oligonucleotide bacterial tiling microarrays.PLoS One. 2009 Jun 17;4(6):e5943. doi: 10.1371/journal.pone.0005943. PLoS One. 2009. PMID: 19536279 Free PMC article.
References
-
- Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. - PubMed
-
- Bloomfield VA, et al. Nucleic Acids: Structures, Properties, and Functions. University Science Books; Herndon, VA, USA: 2000.
-
- Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

