Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme
- PMID: 17643375
- PMCID: PMC4498903
- DOI: 10.1016/j.molcel.2007.06.027
Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme
Abstract
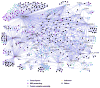
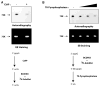
We have performed a survey of soluble human protein complexes containing components of the transcription and RNA processing machineries using protein affinity purification coupled to mass spectrometry. Thirty-two tagged polypeptides yielded a network of 805 high-confidence interactions. Remarkably, the network is significantly enriched in proteins that regulate the formation of protein complexes, including a number of previously uncharacterized proteins for which we have inferred functions. The RNA polymerase II (RNAP II)-associated proteins (RPAPs) are physically and functionally associated with RNAP II, forming an interface between the enzyme and chaperone/scaffolding proteins. BCDIN3 is the 7SK snRNA methylphosphate capping enzyme (MePCE) present in an snRNP complex containing both RNA processing and transcription factors, including the elongation factor P-TEFb. Our results define a high-density protein interaction network for the mammalian transcription machinery and uncover multiple regulatory factors that target the transcription machinery.
Figures




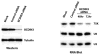
Comment in
-
Transcriptional networking cap-tures the 7SK RNA 5'-gamma-methyltransferase.Mol Cell. 2007 Aug 17;27(4):517-9. doi: 10.1016/j.molcel.2007.08.001. Mol Cell. 2007. PMID: 17707222
Similar articles
-
RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP.Nucleic Acids Res. 2013 Apr;41(8):4686-98. doi: 10.1093/nar/gkt159. Epub 2013 Mar 6. Nucleic Acids Res. 2013. PMID: 23471002 Free PMC article.
-
A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP.Nucleic Acids Res. 2010 Jan;38(2):360-9. doi: 10.1093/nar/gkp977. Epub 2009 Nov 11. Nucleic Acids Res. 2010. PMID: 19906723 Free PMC article.
-
JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.Elife. 2020 Feb 12;9:e53930. doi: 10.7554/eLife.53930. Elife. 2020. PMID: 32048991 Free PMC article.
-
Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb.Nucleic Acids Res. 2016 Sep 19;44(16):7527-39. doi: 10.1093/nar/gkw585. Epub 2016 Jul 1. Nucleic Acids Res. 2016. PMID: 27369380 Free PMC article. Review.
-
Transcription elongation control by the 7SK snRNP complex: Releasing the pause.Cell Cycle. 2016 Aug 17;15(16):2115-2123. doi: 10.1080/15384101.2016.1181241. Epub 2016 May 6. Cell Cycle. 2016. PMID: 27152730 Free PMC article. Review.
Cited by
-
7SKiing on chromatin: Move globally, act locally.RNA Biol. 2016 Jun 2;13(6):545-53. doi: 10.1080/15476286.2016.1181254. Epub 2016 Apr 29. RNA Biol. 2016. PMID: 27128603 Free PMC article. Review.
-
Systematic Determination of Human Cyclin Dependent Kinase (CDK)-9 Interactome Identifies Novel Functions in RNA Splicing Mediated by the DEAD Box (DDX)-5/17 RNA Helicases.Mol Cell Proteomics. 2015 Oct;14(10):2701-21. doi: 10.1074/mcp.M115.049221. Epub 2015 Jul 24. Mol Cell Proteomics. 2015. PMID: 26209609 Free PMC article.
-
Reptin/Ruvbl2 is a Lrrc6/Seahorse interactor essential for cilia motility.Proc Natl Acad Sci U S A. 2013 Jul 30;110(31):12697-702. doi: 10.1073/pnas.1300968110. Epub 2013 Jul 15. Proc Natl Acad Sci U S A. 2013. PMID: 23858445 Free PMC article.
-
7SK small nuclear RNA transcription level down-regulates in human tumors and stem cells.Med Oncol. 2016 Nov;33(11):128. doi: 10.1007/s12032-016-0841-x. Epub 2016 Oct 17. Med Oncol. 2016. PMID: 27752877
-
Maturation and Assembly of mTOR Complexes by the HSP90-R2TP-TTT Chaperone System: Molecular Insights and Mechanisms.Subcell Biochem. 2024;104:459-483. doi: 10.1007/978-3-031-58843-3_17. Subcell Biochem. 2024. PMID: 38963496 Review.
References
-
- Aguilera A. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr Opin Cell Biol. 2005;17:242–250. - PubMed
-
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

