A general strategy to solve the phase problem in RNA crystallography
- PMID: 17637337
- PMCID: PMC1995091
- DOI: 10.1016/j.str.2007.06.003
A general strategy to solve the phase problem in RNA crystallography
Abstract
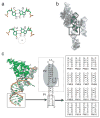
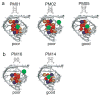
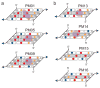
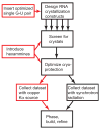
X-ray crystallography of biologically important RNA molecules has been hampered by technical challenges, including finding heavy-atom derivatives to obtain high-quality experimental phase information. Existing techniques have drawbacks, limiting the rate at which important new structures are solved. To address this, we have developed a reliable means to localize heavy atoms specifically to virtually any RNA. By solving the crystal structures of thirteen variants of the G*U wobble pair cation binding motif, we have identified a version that when inserted into an RNA helix introduces a high-occupancy cation binding site suitable for phasing. This "directed soaking" strategy can be integrated fully into existing RNA crystallography methods, potentially increasing the rate at which important structures are solved and facilitating routine solving of structures using Cu-Kalpha radiation. This method already has been used to solve several crystal structures.
Figures

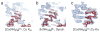
Similar articles
-
Soaking Hexammine Cations into RNA Crystals to Obtain Derivatives for Phasing Diffraction Data.Methods Mol Biol. 2016;1320:219-32. doi: 10.1007/978-1-4939-2763-0_14. Methods Mol Biol. 2016. PMID: 26227046 Free PMC article.
-
Crystal structure of the ribonucleoprotein core of the signal recognition particle.Science. 2000 Feb 18;287(5456):1232-9. doi: 10.1126/science.287.5456.1232. Science. 2000. PMID: 10678824
-
A surprising function for SRP RNA?Nat Struct Biol. 2000 Mar;7(3):179-81. doi: 10.1038/73265. Nat Struct Biol. 2000. PMID: 10700269 No abstract available.
-
Selenium derivatization of nucleic acids for X-ray crystal-structure and function studies.Chem Biodivers. 2010 Apr;7(4):753-85. doi: 10.1002/cbdv.200900200. Chem Biodivers. 2010. PMID: 20397215 Review.
-
Nucleic acid X-ray crystallography via direct selenium derivatization.Chem Soc Rev. 2011 Sep;40(9):4591-602. doi: 10.1039/c1cs15020k. Epub 2011 Jun 13. Chem Soc Rev. 2011. PMID: 21666919 Review.
Cited by
-
Structural-profiling of low molecular weight RNAs by nanopore trapping/translocation using Mycobacterium smegmatis porin A.Nat Commun. 2021 Jun 7;12(1):3368. doi: 10.1038/s41467-021-23764-y. Nat Commun. 2021. PMID: 34099723 Free PMC article.
-
Enhanced group II intron retrohoming in magnesium-deficient Escherichia coli via selection of mutations in the ribozyme core.Proc Natl Acad Sci U S A. 2013 Oct 1;110(40):E3800-9. doi: 10.1073/pnas.1315742110. Epub 2013 Sep 16. Proc Natl Acad Sci U S A. 2013. PMID: 24043808 Free PMC article.
-
The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer.Structure. 2011 Oct 12;19(10):1413-23. doi: 10.1016/j.str.2011.06.019. Epub 2011 Sep 8. Structure. 2011. PMID: 21906956 Free PMC article.
-
New molecular engineering approaches for crystallographic studies of large RNAs.Curr Opin Struct Biol. 2014 Jun;26:9-15. doi: 10.1016/j.sbi.2014.02.001. Epub 2014 Mar 6. Curr Opin Struct Biol. 2014. PMID: 24607443 Free PMC article. Review.
-
sincFold: end-to-end learning of short- and long-range interactions in RNA secondary structure.Brief Bioinform. 2024 May 23;25(4):bbae271. doi: 10.1093/bib/bbae271. Brief Bioinform. 2024. PMID: 38855913 Free PMC article.
References
-
- Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2004;11:53–55. - PubMed
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
-
- Batey RT, Doudna JA. Structural and energetic analysis of metal ions essential to SRP signal recognition domain assembly. Biochemistry. 2002;41:11703–11710. - PubMed
-
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. - PubMed
-
- Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

