Mammalian heparanase: what is the message?
- PMID: 17635638
- PMCID: PMC3922351
- DOI: 10.1111/j.1582-4934.2007.00039.x
Mammalian heparanase: what is the message?
Abstract
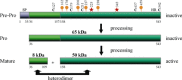
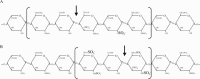
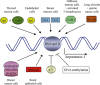
Heparan sulphate proteoglycans are ubiquitous macromolecules of cell surfaces and extracellular matrices. Numerous extracellular matrix proteins, growth factors, morphogens, cytokines, chemokines and coagulation factors are bound and regulated by heparan sulphate. Degradation of heparan sulphate thus potentially profoundly affects cell and tissue function. Although there is evidence that several heparan sulphate-degrading endoglucuronidases (heparanases) might exist, so far only one transcript encoding a functional heparanase has been identified: heparanase-1. In the first part of this review, we discuss the current knowledge about heparan sulphate proteoglycans and the functional importance of their versatile interactions. In the second part, we summarize recent findings that have contributed to the characterization of heparanase-1, focusing on the molecular properties, working mechanism, substrate specificity, expression pattern, cellular activation and localization of this enzyme. Additionally, we review data implicating heparanase-1 in several normal and pathological processes, focusing on tumour metastasis and angiogenesis, and on evidence for a potentially direct signalling function of the molecule. In that context, we also briefly discuss heparanase-2, an intriguing close homologue of heparanase-1, for which, so far, no heparan sulphate-degrading activity could be demonstrated.
Figures


Similar articles
-
Mammalian heparanase: involvement in cancer metastasis, angiogenesis and normal development.Semin Cancer Biol. 2002 Apr;12(2):121-9. doi: 10.1006/scbi.2001.0420. Semin Cancer Biol. 2002. PMID: 12027584 Review.
-
A functional heparan sulfate mimetic implicates both heparanase and heparan sulfate in tumor angiogenesis and invasion in a mouse model of multistage cancer.Oncogene. 2005 Jun 9;24(25):4037-51. doi: 10.1038/sj.onc.1208602. Oncogene. 2005. PMID: 15806157
-
Heparanase: Historical Aspects and Future Perspectives.Adv Exp Med Biol. 2020;1221:71-96. doi: 10.1007/978-3-030-34521-1_3. Adv Exp Med Biol. 2020. PMID: 32274707 Review.
-
[Research advancement on heparanase in tumor metastasis].Ai Zheng. 2005 Sep;24(9):1156-60. Ai Zheng. 2005. PMID: 16159446 Review. Chinese.
-
Heparanase: one molecule with multiple functions in cancer progression.Connect Tissue Res. 2008;49(3):207-10. doi: 10.1080/03008200802143281. Connect Tissue Res. 2008. PMID: 18661344 Review.
Cited by
-
The prognostic significance of heparanase expression in metastatic melanoma.Oncotarget. 2016 Nov 15;7(46):74678-74685. doi: 10.18632/oncotarget.12492. Oncotarget. 2016. PMID: 27732945 Free PMC article.
-
Heparanase upregulates Th2 cytokines, ameliorating experimental autoimmune encephalitis.Mol Immunol. 2010 Jun;47(10):1890-8. doi: 10.1016/j.molimm.2010.03.014. Mol Immunol. 2010. PMID: 20399501 Free PMC article.
-
Heparanase and Chemotherapy Synergize to Drive Macrophage Activation and Enhance Tumor Growth.Cancer Res. 2020 Jan 1;80(1):57-68. doi: 10.1158/0008-5472.CAN-19-1676. Epub 2019 Nov 5. Cancer Res. 2020. PMID: 31690669 Free PMC article.
-
Proteoglycans in health and disease: new concepts for heparanase function in tumor progression and metastasis.FEBS J. 2010 Oct;277(19):3890-903. doi: 10.1111/j.1742-4658.2010.07799.x. Epub 2010 Aug 31. FEBS J. 2010. PMID: 20840586 Free PMC article. Review.
-
Heparanase in inflammation and inflammation-associated cancer.FEBS J. 2013 May;280(10):2307-19. doi: 10.1111/febs.12184. Epub 2013 Mar 4. FEBS J. 2013. PMID: 23398975 Free PMC article. Review.
References
-
- Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–77. - PubMed
-
- Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–71. - PubMed
-
- Chen RL, Lander AD. Mechanisms underlying preferential assembly of heparan sulfate on glypican-1. J Biol Chem. 2001;276:7507–17. - PubMed
-
- Esko JD, Zhang L. Influence of core protein sequence on glycosaminoglycan assembly. Curr Opin Struct Biol. 1996;6:663–70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

