Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication
- PMID: 17634238
- PMCID: PMC2045455
- DOI: 10.1128/JVI.00017-07
Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication
Abstract
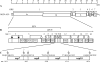
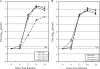
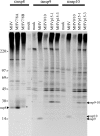
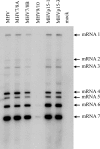
Coronaviruses express open reading frame 1a (ORF1a) and ORF1b polyproteins from which 16 nonstructural proteins (nsp) are derived. The highly conserved region at the carboxy terminus of ORF1a is processed by the nsp5 proteinase (Mpro) into mature products, including nsp7, nsp8, nsp9, and nsp10, proteins with predicted or identified activities involved in RNA synthesis. Although continuous translation and proteolytic processing of ORF1ab by Mpro is required for replication, it is unknown whether specific cleavage events within the polyprotein are dispensable. We determined the requirement for the nsp7 to nsp10 proteins and their processing during murine hepatitis virus (MHV) replication. Through use of an MHV reverse genetics system, in-frame deletions of the coding sequences for nsp7 to nsp10, or ablation of their flanking Mpro cleavage sites, were made and the effects upon replication were determined. Viable viruses were characterized by analysis of Mpro processing, RNA transcription, and growth fitness. Deletion of any of the regions encoding nsp7 to nsp10 was lethal. Disruption of the cleavage sites was lethal with the exception of that of the nsp9-nsp10 site, which resulted in a mutant virus with attenuated replication. Passage of the attenuated nsp9-nsp10 cleavage mutant increased fitness to near-wild-type kinetics without reversion to a virus capable of processing nsp9-nsp10. We also confirmed the presence of a second cleavage site between nsp7 and nsp8. In order to determine whether a distinct function could be attributed to preprocessed forms of the polyprotein, including nsp7 to nsp10, the genes encoding nsp7 and nsp8 were rearranged. The mutant virus was not viable, suggesting that the uncleaved protein may be essential for replication or proteolytic processing.
Figures


Similar articles
-
MHV-A59 ORF1a replicase protein nsp7-nsp10 processing in replication.Adv Exp Med Biol. 2006;581:101-4. doi: 10.1007/978-0-387-33012-9_17. Adv Exp Med Biol. 2006. PMID: 17037513 Free PMC article. No abstract available.
-
Proteolytic Processing of the Coronavirus Replicase Nonstructural Protein 14 Exonuclease Is Not Required for Virus Replication but Alters RNA Synthesis and Viral Fitness.J Virol. 2022 Aug 24;96(16):e0084122. doi: 10.1128/jvi.00841-22. Epub 2022 Aug 4. J Virol. 2022. PMID: 35924922 Free PMC article.
-
Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis.J Virol. 2007 Jun;81(12):6356-68. doi: 10.1128/JVI.02805-06. Epub 2007 Mar 28. J Virol. 2007. PMID: 17392363 Free PMC article.
-
The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins.Virus Res. 2010 Dec;154(1-2):61-76. doi: 10.1016/j.virusres.2010.07.030. Epub 2010 Aug 7. Virus Res. 2010. PMID: 20696193 Free PMC article. Review.
-
The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing.Adv Virus Res. 2016;96:59-126. doi: 10.1016/bs.aivir.2016.08.008. Epub 2016 Sep 14. Adv Virus Res. 2016. PMID: 27712628 Free PMC article. Review.
Cited by
-
SARS-CoV-2 RNA-dependent RNA polymerase as a target for high-throughput drug screening.Future Virol. 2022 Jan:10.2217/fvl-2021-0335. doi: 10.2217/fvl-2021-0335. Epub 2023 Feb 3. Future Virol. 2022. PMID: 36794167 Free PMC article. Review.
-
Selenoprotein S Interacts with the Replication and Transcription Complex of SARS-CoV-2 by Binding nsp7.J Mol Biol. 2023 Apr 15;435(8):168008. doi: 10.1016/j.jmb.2023.168008. Epub 2023 Feb 10. J Mol Biol. 2023. PMID: 36773692 Free PMC article.
-
Synthesis and In Silico Investigation of Isatin-Based Schiff Bases as Potential Inhibitors for Promising Targets against SARS-CoV-2.ChemistrySelect. 2022 Dec 13;7(46):e202201983. doi: 10.1002/slct.202201983. Epub 2022 Dec 9. ChemistrySelect. 2022. PMID: 36718466 Free PMC article.
-
Severe acute respiratory syndrome coronaviruses contributing to mitochondrial dysfunction: Implications for post-COVID complications.Mitochondrion. 2023 Mar;69:43-56. doi: 10.1016/j.mito.2023.01.005. Epub 2023 Jan 20. Mitochondrion. 2023. PMID: 36690315 Free PMC article. Review.
-
Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.Nucleic Acids Res. 2023 Jan 11;51(1):475-487. doi: 10.1093/nar/gkac1207. Nucleic Acids Res. 2023. PMID: 36546776 Free PMC article.
References
-
- Bonilla, P. J., S. A. Hughes, J. D. Pinon, and S. R. Weiss. 1995. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology 209:489-497. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

