Overlapping roles of the Rous sarcoma virus Gag p10 domain in nuclear export and virion core morphology
- PMID: 17634229
- PMCID: PMC2045444
- DOI: 10.1128/JVI.01061-07
Overlapping roles of the Rous sarcoma virus Gag p10 domain in nuclear export and virion core morphology
Abstract
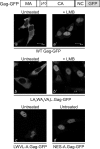
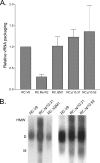
Nucleocytoplasmic shuttling of the Rous sarcoma virus (RSV) Gag polyprotein is an integral step in virus particle assembly. A nuclear export signal (NES) was previously identified within the p10 domain of RSV Gag. Gag mutants containing deletions of the p10 NES or mutations of critical hydrophobic residues at positions 219, 222, 225, or 229 become trapped within the nucleus and exhibit defects in the efficiency of virus particle release. To investigate other potential roles for Gag nuclear trafficking in RSV replication, we created viruses bearing NES mutant Gag proteins. Viruses carrying p10 mutations produced low levels of particles, as anticipated, and those particles that were released were noninfectious. The p10 mutant viruses contained approximately normal amounts of Gag, Gag-Pol, and Env proteins and genomic viral RNA (vRNA), but several major structural defects were found. Thin-section transmission electron microscopy revealed that the mature particles appeared misshapen, while the viral cores were cylindrical, horseshoe-shaped, or fragmented, with some particles containing multiple small, electron-dense aggregates. Immature virus-like particles produced by the expression of Gag proteins bearing p10 mutations were also aberrant, with both spherical and tubular filamentous particles produced. Interestingly, the secondary structure of the encapsidated vRNA was altered; although dimeric vRNA was predominant, there was an additional high-molecular-weight fraction. Together, these results indicate that the p10 NES domain of Gag is critical for virus replication and that it plays overlapping roles required for the nuclear shuttling of Gag and for the maintenance of proper virion core morphology.
Figures






Similar articles
-
Visualizing Association of the Retroviral Gag Protein with Unspliced Viral RNA in the Nucleus.mBio. 2020 Apr 7;11(2):e00524-20. doi: 10.1128/mBio.00524-20. mBio. 2020. PMID: 32265329 Free PMC article.
-
Role of the Rous sarcoma virus p10 domain in shape determination of gag virus-like particles assembled in vitro and within Escherichia coli.J Virol. 2000 Nov;74(21):10260-8. doi: 10.1128/jvi.74.21.10260-10268.2000. J Virol. 2000. PMID: 11024160 Free PMC article.
-
Insertion of a classical nuclear import signal into the matrix domain of the Rous sarcoma virus Gag protein interferes with virus replication.J Virol. 2004 Dec;78(24):13534-42. doi: 10.1128/JVI.78.24.13534-13542.2004. J Virol. 2004. PMID: 15564464 Free PMC article.
-
The choreography of HIV-1 proteolytic processing and virion assembly.J Biol Chem. 2012 Nov 30;287(49):40867-74. doi: 10.1074/jbc.R112.399444. Epub 2012 Oct 5. J Biol Chem. 2012. PMID: 23043111 Free PMC article. Review.
-
Synthesis and assembly of chimeric human immunodeficiency virus gag pseudovirions.Intervirology. 1996;39(1-2):40-8. doi: 10.1159/000150473. Intervirology. 1996. PMID: 8957668 Review.
Cited by
-
A nuclear export signal within the structural Gag protein is required for prototype foamy virus replication.Retrovirology. 2011 Jan 21;8(1):6. doi: 10.1186/1742-4690-8-6. Retrovirology. 2011. PMID: 21255441 Free PMC article.
-
HIV-1 Gag co-localizes with euchromatin histone marks at the nuclear periphery.J Virol. 2023 Dec 21;97(12):e0117923. doi: 10.1128/jvi.01179-23. Epub 2023 Nov 22. J Virol. 2023. PMID: 37991367 Free PMC article.
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.Retrovirology. 2024 Jun 19;21(1):13. doi: 10.1186/s12977-024-00645-y. Retrovirology. 2024. PMID: 38898526 Free PMC article.
-
Molecular events accompanying rous sarcoma virus rescue from rodent cells and the role of viral gene complementation.J Virol. 2014 Mar;88(6):3505-15. doi: 10.1128/JVI.02761-13. Epub 2014 Jan 8. J Virol. 2014. PMID: 24403579 Free PMC article.
-
Functional Equivalence of Retroviral MA Domains in Facilitating Psi RNA Binding Specificity by Gag.Viruses. 2016 Sep 19;8(9):256. doi: 10.3390/v8090256. Viruses. 2016. PMID: 27657107 Free PMC article.
References
-
- Bourmakina, S. V., and A. Garcia-Sastre. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517-527. - PubMed
-
- Briggs, J. A., M. C. Johnson, M. N. Simon, S. D. Fuller, and V. M. Vogt. 2006. Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J. Mol. Biol. 355:157-168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

