Binding parameters and thermodynamics of the interaction of the human cytomegalovirus DNA polymerase accessory protein, UL44, with DNA: implications for the processivity mechanism
- PMID: 17617644
- PMCID: PMC1950537
- DOI: 10.1093/nar/gkm506
Binding parameters and thermodynamics of the interaction of the human cytomegalovirus DNA polymerase accessory protein, UL44, with DNA: implications for the processivity mechanism
Abstract
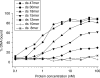
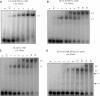
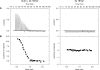
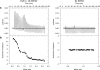
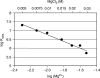
The mechanisms of processivity factors of herpesvirus DNA polymerases remain poorly understood. The proposed processivity factor for human cytomegalovirus DNA polymerase is a DNA-binding protein, UL44. Previous findings, including the crystal structure of UL44, have led to the hypothesis that UL44 binds DNA as a dimer via lysine residues. To understand how UL44 interacts with DNA, we used filter-binding and electrophoretic mobility shift assays and isothermal titration calorimetry (ITC) analysis of binding to oligonucleotides. UL44 bound directly to double-stranded DNA as short as 12 bp, with apparent dissociation constants in the nanomolar range for DNAs >18 bp, suggesting a minimum DNA length for UL44 interaction. UL44 also bound single-stranded DNA, albeit with lower affinity, and for either single- or double-stranded DNA, there was no apparent sequence specificity. ITC analysis revealed that UL44 binds to duplex DNA as a dimer. Binding was endothermic, indicating an entropically driven process, likely due to release of bound ions. Consistent with this hypothesis, analysis of the relationship between binding and ionic strength indicated that, on average, 4 +/- 1 monovalent ions are released in the interaction of each monomer of UL44 with DNA. The results taken together reveal interesting implications for how UL44 may mediate processivity.
Figures
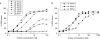

Similar articles
-
Specific residues in the connector loop of the human cytomegalovirus DNA polymerase accessory protein UL44 are crucial for interaction with the UL54 catalytic subunit.J Virol. 2004 Sep;78(17):9084-92. doi: 10.1128/JVI.78.17.9084-9092.2004. J Virol. 2004. PMID: 15308704 Free PMC article.
-
The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer.Mol Cell. 2004 Jul 23;15(2):233-44. doi: 10.1016/j.molcel.2004.06.018. Mol Cell. 2004. PMID: 15260974
-
Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44.J Biol Chem. 2006 Feb 24;281(8):5224-32. doi: 10.1074/jbc.M506900200. Epub 2005 Dec 20. J Biol Chem. 2006. PMID: 16371349
-
Advances in the analysis of isothermal titration calorimetry data for ligand-DNA interactions.Methods. 2007 Jun;42(2):162-72. doi: 10.1016/j.ymeth.2007.01.010. Methods. 2007. PMID: 17472898 Review.
-
How do DNA-bound proteins leave their binding sites? The role of facilitated dissociation.Curr Opin Chem Biol. 2019 Dec;53:118-124. doi: 10.1016/j.cbpa.2019.08.007. Epub 2019 Oct 2. Curr Opin Chem Biol. 2019. PMID: 31586479 Free PMC article. Review.
Cited by
-
The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner.PLoS One. 2012;7(11):e49630. doi: 10.1371/journal.pone.0049630. Epub 2012 Nov 15. PLoS One. 2012. PMID: 23166733 Free PMC article.
-
The positively charged surface of herpes simplex virus UL42 mediates DNA binding.J Biol Chem. 2008 Mar 7;283(10):6154-61. doi: 10.1074/jbc.M708691200. Epub 2008 Jan 4. J Biol Chem. 2008. PMID: 18178550 Free PMC article.
-
Role of homodimerization of human cytomegalovirus DNA polymerase accessory protein UL44 in origin-dependent DNA replication in cells.J Virol. 2008 Dec;82(24):12574-9. doi: 10.1128/JVI.01193-08. Epub 2008 Oct 8. J Virol. 2008. PMID: 18842734 Free PMC article.
-
Nucleolin associates with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for efficient viral replication.J Virol. 2010 Feb;84(4):1771-84. doi: 10.1128/JVI.01510-09. Epub 2009 Dec 9. J Virol. 2010. PMID: 20007282 Free PMC article.
-
The 6-aminoquinolone WC5 inhibits human cytomegalovirus replication at an early stage by interfering with the transactivating activity of viral immediate-early 2 protein.Antimicrob Agents Chemother. 2010 May;54(5):1930-40. doi: 10.1128/AAC.01730-09. Epub 2010 Mar 1. Antimicrob Agents Chemother. 2010. PMID: 20194695 Free PMC article.
References
-
- Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. - PubMed
-
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. - PubMed
-
- Jeruzalmi D, O’Donnell M, Kuriyan J. Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol. 2002;12:217–224. - PubMed
-
- Randell JC, Coen DM. The herpes simplex virus processivity factor, UL42, binds DNA as a monomer. J. Mol. Biol. 2004;335:409–413. - PubMed

