OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria
- PMID: 17615298
- PMCID: PMC1951777
- DOI: 10.1091/mbc.e07-02-0164
OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria
Abstract
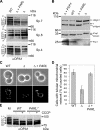
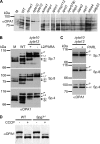
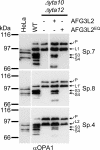
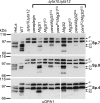
The morphology of mitochondria in mammalian cells is regulated by proteolytic cleavage of OPA1, a dynamin-like GTPase of the mitochondrial inner membrane. The mitochondrial rhomboid protease PARL, and paraplegin, a subunit of the ATP-dependent m-AAA protease, were proposed to be involved in this process. Here, we characterized individual OPA1 isoforms by mass spectrometry, and we reconstituted their processing in yeast to identify proteases involved in OPA1 cleavage. The yeast homologue of OPA1, Mgm1, was processed both by PARL and its yeast homologue Pcp1. Neither of these rhomboid proteases cleaved OPA1. The formation of small OPA1 isoforms was impaired in yeast cells lacking the m-AAA protease subunits Yta10 and Yta12 and was restored upon expression of murine or human m-AAA proteases. OPA1 processing depended on the subunit composition of mammalian m-AAA proteases. Homo-oligomeric m-AAA protease complexes composed of murine Afg3l1, Afg3l2, or human AFG3L2 subunits cleaved OPA1 with higher efficiency than paraplegin-containing m-AAA proteases. OPA1 processing proceeded normally in murine cell lines lacking paraplegin or PARL. Our results provide evidence for different substrate specificities of m-AAA proteases composed of different subunits and reveal a striking evolutionary switch of proteases involved in the proteolytic processing of dynamin-like GTPases in mitochondria.
Figures

Similar articles
-
Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1.J Cell Biol. 2009 Dec 28;187(7):1023-36. doi: 10.1083/jcb.200906084. J Cell Biol. 2009. PMID: 20038678 Free PMC article.
-
Variable and tissue-specific subunit composition of mitochondrial m-AAA protease complexes linked to hereditary spastic paraplegia.Mol Cell Biol. 2007 Jan;27(2):758-67. doi: 10.1128/MCB.01470-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101804 Free PMC article.
-
Autocatalytic processing of m-AAA protease subunits in mitochondria.Mol Biol Cell. 2009 Oct;20(19):4216-24. doi: 10.1091/mbc.e09-03-0218. Epub 2009 Aug 5. Mol Biol Cell. 2009. PMID: 19656850 Free PMC article.
-
m-AAA proteases, mitochondrial calcium homeostasis and neurodegeneration.Cell Res. 2018 Mar;28(3):296-306. doi: 10.1038/cr.2018.17. Epub 2018 Feb 16. Cell Res. 2018. PMID: 29451229 Free PMC article. Review.
-
ATP-dependent proteases controlling mitochondrial function in the yeast Saccharomyces cerevisiae.Cell Mol Life Sci. 1999 Nov 30;56(9-10):825-42. doi: 10.1007/s000180050029. Cell Mol Life Sci. 1999. PMID: 11212342 Free PMC article. Review.
Cited by
-
MIC26 and MIC27 cooperate to regulate cardiolipin levels and the landscape of OXPHOS complexes.Life Sci Alliance. 2020 Aug 11;3(10):e202000711. doi: 10.26508/lsa.202000711. Print 2020 Oct. Life Sci Alliance. 2020. PMID: 32788226 Free PMC article.
-
Importing mitochondrial proteins: machineries and mechanisms.Cell. 2009 Aug 21;138(4):628-44. doi: 10.1016/j.cell.2009.08.005. Cell. 2009. PMID: 19703392 Free PMC article. Review.
-
Mitochondrial quality control: a matter of life and death for neurons.EMBO J. 2012 Mar 21;31(6):1336-49. doi: 10.1038/emboj.2012.38. Epub 2012 Feb 21. EMBO J. 2012. PMID: 22354038 Free PMC article. Review.
-
Rhomboid protease PARL mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5.J Biol Chem. 2012 Oct 5;287(41):34635-45. doi: 10.1074/jbc.M112.357509. Epub 2012 Aug 22. J Biol Chem. 2012. PMID: 22915595 Free PMC article.
-
OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution.Genome Res. 2011 Jan;21(1):12-20. doi: 10.1101/gr.108696.110. Epub 2010 Oct 25. Genome Res. 2011. PMID: 20974897 Free PMC article.
References
-
- Alexander C., et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000;26:211–215. - PubMed
-
- Arnoult D., Grodet A., Lee Y. J., Estaquier J., Blackstone C. Release of OPA1 during apoptosis participates in the rapid and complete release of cytochrome c and subsequent mitochondrial fragmentation. J. Biol. Chem. 2005;280:35742–35750. - PubMed
-
- Casari G., et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

