Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance
- PMID: 17600134
- PMCID: PMC1949876
- DOI: 10.1104/pp.107.103762
Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance
Abstract
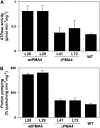
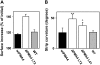
The plasma membrane proton pump ATPase (H(+)-ATPase) plays a major role in the activation of ion and nutrient transport and has been suggested to be involved in several physiological processes, such as cell expansion and salt tolerance. Its activity is regulated by a C-terminal autoinhibitory domain that can be displaced by phosphorylation and the binding of regulatory 14-3-3 proteins, resulting in an activated enzyme. To better understand the physiological consequence of this activation, we have analyzed transgenic tobacco (Nicotiana tabacum) plants expressing either wild-type plasma membrane H(+)-ATPase4 (wtPMA4) or a PMA4 mutant lacking the autoinhibitory domain (DeltaPMA4), generating a constitutively activated enzyme. Plants showing 4-fold higher expression of wtPMA4 than untransformed plants did not display any unusual phenotype and their leaf and root external acidification rates were not modified, while their in vitro H(+)-ATPase activity was markedly increased. This indicates that, in vivo, H(+)-ATPase overexpression is compensated by down-regulation of H(+)-ATPase activity. In contrast, plants that expressed DeltaPMA4 were characterized by a lower apoplastic and external root pH, abnormal leaf inclination, and twisted stems, suggesting alterations in cell expansion. This was confirmed by in vitro leaf extension and curling assays. These data therefore strongly support a direct role of H(+)-ATPase in plant development. The DeltaPMA4 plants also displayed increased salt tolerance during germination and seedling growth, supporting the hypothesis that H(+)-ATPase is involved in salt tolerance.
Figures



Similar articles
-
Two widely expressed plasma membrane H(+)-ATPase isoforms of Nicotiana tabacum are differentially regulated by phosphorylation of their penultimate threonine.Plant J. 2010 Apr;62(2):291-301. doi: 10.1111/j.1365-313X.2010.04147.x. Epub 2010 Jan 27. Plant J. 2010. PMID: 20128881
-
SpAHA1 and SpSOS1 Coordinate in Transgenic Yeast to Improve Salt Tolerance.PLoS One. 2015 Sep 4;10(9):e0137447. doi: 10.1371/journal.pone.0137447. eCollection 2015. PLoS One. 2015. PMID: 26340746 Free PMC article.
-
Over-expression of a plasma membrane H+-ATPase SpAHA1 conferred salt tolerance to transgenic Arabidopsis.Protoplasma. 2018 Nov;255(6):1827-1837. doi: 10.1007/s00709-018-1275-4. Epub 2018 Jun 13. Protoplasma. 2018. PMID: 29948367
-
The plasma membrane proton pump ATPase: the significance of gene subfamilies.Planta. 2003 Jan;216(3):355-65. doi: 10.1007/s00425-002-0856-8. Epub 2002 Sep 6. Planta. 2003. PMID: 12520326 Review.
-
Regulation of plasma membrane H(+)-ATPase in fungi and plants.Biochim Biophys Acta. 2000 Mar 10;1469(1):31-42. doi: 10.1016/s0304-4157(99)00011-8. Biochim Biophys Acta. 2000. PMID: 10692636 Review.
Cited by
-
Arabidopsis COP1 guides stomatal response in guard cells through pH regulation.Commun Biol. 2024 Feb 5;7(1):150. doi: 10.1038/s42003-024-05847-w. Commun Biol. 2024. PMID: 38316905 Free PMC article.
-
Expression of a constitutively activated plasma membrane H+-ATPase in Nicotiana tabacum BY-2 cells results in cell expansion.Planta. 2016 Nov;244(5):1109-1124. doi: 10.1007/s00425-016-2571-x. Epub 2016 Jul 21. Planta. 2016. PMID: 27444008
-
Identification and Characterization of miRNAs and lncRNAs Associated with Salinity Stress in Rice Panicles.Int J Mol Sci. 2024 Jul 28;25(15):8247. doi: 10.3390/ijms25158247. Int J Mol Sci. 2024. PMID: 39125819 Free PMC article.
-
Analysis of tomato plasma membrane H(+)-ATPase gene family suggests a mycorrhiza-mediated regulatory mechanism conserved in diverse plant species.Mycorrhiza. 2016 Oct;26(7):645-56. doi: 10.1007/s00572-016-0700-9. Epub 2016 Apr 22. Mycorrhiza. 2016. PMID: 27103309
-
The root apoplastic pH as an integrator of plant signaling.Front Plant Sci. 2022 Aug 23;13:931979. doi: 10.3389/fpls.2022.931979. eCollection 2022. Front Plant Sci. 2022. PMID: 36082302 Free PMC article. Review.
References
-
- Alexander MP (1980) A versatile stain for pollen, fungi, yeast and bacteria. Stain Technol 55 13–18 - PubMed
-
- Amborabé BE, Fleurat-Lessard P, Bonmort J, Roustan JP, Roblin G (2001) Effects of eutypine, a toxin from Eutypa lata, on plant cell plasma membrane: possible subsequent implication in disease development. Plant Physiol Biochem 39 51–58
-
- Arango M, Gévaudant F, Oufattole M, Boutry M (2003) The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216 355–365 - PubMed
-
- Baunsgaard L, Fuglsang AT, Jahn T, Korthout HAAJ, de Boer AH, Palmgren MG (1998) The 14-3-3 proteins associate with the plant plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J 13 661–671 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

