Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance
- PMID: 17600086
- PMCID: PMC1933516
- DOI: 10.1101/gr.6468307
Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance
Abstract
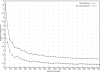
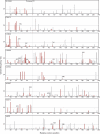
The detection of mutant spectra within a population of microorganisms is critical for the management of drug-resistant infections. We performed ultra-deep pyrosequencing to detect minor sequence variants in HIV-1 protease and reverse transcriptase (RT) genes from clinical plasma samples. We estimated empirical error rates from four HIV-1 plasmid clones and used them to develop a statistical approach to distinguish authentic minor variants from sequencing errors in eight clinical samples. Ultra-deep pyrosequencing detected an average of 58 variants per sample compared with an average of eight variants per sample detected by conventional direct-PCR dideoxynucleotide sequencing. In the clinical sample with the largest number of minor sequence variants, all 60 variants present in > or =3% of genomes and 20 of 35 variants present in <3% of genomes were confirmed by limiting dilution sequencing. With appropriate analysis, ultra-deep pyrosequencing is a promising method for characterizing genetic diversity and detecting minor yet clinically relevant variants in biological samples with complex genetic populations.
Figures
Similar articles
-
HIV-1 Protease, Reverse Transcriptase, and Integrase Variation.J Virol. 2016 Jun 10;90(13):6058-6070. doi: 10.1128/JVI.00495-16. Print 2016 Jul 1. J Virol. 2016. PMID: 27099321 Free PMC article.
-
Evaluation of different analysis pipelines for the detection of HIV-1 minority resistant variants.PLoS One. 2018 Jun 1;13(6):e0198334. doi: 10.1371/journal.pone.0198334. eCollection 2018. PLoS One. 2018. PMID: 29856864 Free PMC article.
-
HIV drug resistance surveillance using pooled pyrosequencing.PLoS One. 2010 Feb 17;5(2):e9263. doi: 10.1371/journal.pone.0009263. PLoS One. 2010. PMID: 20174661 Free PMC article.
-
Update of the drug resistance mutations in HIV-1: 2004.Top HIV Med. 2004 Oct-Nov;12(4):119-24. Top HIV Med. 2004. PMID: 15516709 Review. No abstract available.
-
HIV sequence databases.AIDS Rev. 2003 Jan-Mar;5(1):52-61. AIDS Rev. 2003. PMID: 12875108 Free PMC article. Review.
Cited by
-
Reverse genetics in high throughput: rapid generation of complete negative strand RNA virus cDNA clones and recombinant viruses thereof.Sci Rep. 2016 Apr 5;6:23887. doi: 10.1038/srep23887. Sci Rep. 2016. PMID: 27046474 Free PMC article.
-
Restriction of V3 region sequence divergence in the HIV-1 envelope gene during antiretroviral treatment in a cohort of recent seroconverters.Retrovirology. 2013 Jan 18;10:8. doi: 10.1186/1742-4690-10-8. Retrovirology. 2013. PMID: 23331949 Free PMC article. Clinical Trial.
-
Application of next-generation sequencing technologies in virology.J Gen Virol. 2012 Sep;93(Pt 9):1853-1868. doi: 10.1099/vir.0.043182-0. Epub 2012 May 30. J Gen Virol. 2012. PMID: 22647373 Free PMC article. Review.
-
Whole-genome sequencing of the efficient industrial fuel-ethanol fermentative Saccharomyces cerevisiae strain CAT-1.Mol Genet Genomics. 2012 Jun;287(6):485-94. doi: 10.1007/s00438-012-0695-7. Epub 2012 May 6. Mol Genet Genomics. 2012. PMID: 22562254
-
HMM-FRAME: accurate protein domain classification for metagenomic sequences containing frameshift errors.BMC Bioinformatics. 2011 May 24;12:198. doi: 10.1186/1471-2105-12-198. BMC Bioinformatics. 2011. PMID: 21609463 Free PMC article.
References
-
- Cai F., Chen H., Hicks C.B., Bartlett J.A., Zhu J., Gao F., Chen H., Hicks C.B., Bartlett J.A., Zhu J., Gao F., Hicks C.B., Bartlett J.A., Zhu J., Gao F., Bartlett J.A., Zhu J., Gao F., Zhu J., Gao F., Gao F. Detection of minor drug-resistant populations by parallel allele-specific sequencing. Nat. Methods. 2007;4:123–125. - PubMed
-
- Coffin J.M. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. - PubMed
-
- Edelstein R.E., Nickerson D.A., Tobe V.O., Manns-Arcuino L.A., Frenkel L.M., Nickerson D.A., Tobe V.O., Manns-Arcuino L.A., Frenkel L.M., Tobe V.O., Manns-Arcuino L.A., Frenkel L.M., Manns-Arcuino L.A., Frenkel L.M., Frenkel L.M. Oligonucleotide ligation assay for detecting mutations in the human immunodeficiency virus type 1 pol gene that are associated with resistance to zidovudine, didanosine, and lamivudine. J. Clin. Microbiol. 1998;36:569–572. - PMC - PubMed
-
- Ellis G.M., Mahalanabis M., Beck I.A., Pepper G., Wright A., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Mahalanabis M., Beck I.A., Pepper G., Wright A., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Beck I.A., Pepper G., Wright A., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Pepper G., Wright A., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Wright A., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Naugler W.E., Pawluk D.M., Li C.C., Pawluk D.M., Li C.C., Li C.C., et al. Comparison of oligonucleotide ligation assay and consensus sequencing for detection of drug-resistant mutants of human immunodeficiency virus type 1 in peripheral blood mononuclear cells and plasma. J. Clin. Microbiol. 2004;42:3670–3674. - PMC - PubMed
-
- Flys T., Nissley D.V., Claasen C.W., Jones D., Shi C., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Nissley D.V., Claasen C.W., Jones D., Shi C., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Claasen C.W., Jones D., Shi C., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Jones D., Shi C., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Shi C., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Mmiro F., Strathern J.N., Jackson J.B., Strathern J.N., Jackson J.B., Jackson J.B., et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J. Infect. Dis. 2005;192:24–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
