Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes
- PMID: 17599067
- PMCID: PMC1933404
- DOI: 10.1038/sj.emboj.7601773
Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes
Abstract
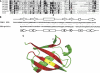
TANK-binding kinase 1 (TBK1/NAK/T2K) and I-kappaB Kinase (IKK-i/IKK-epsilon) play important roles in the regulation of interferon (IFN)-inducible genes during the immune response to bacterial and viral infections. Cell stimulation with ssRNA virus, dsDNA virus or gram-negative bacteria leads to activation of TBK1 or IKK-i, which in turn phosphorylates the transcription factors, IFN-regulatory factor (IRF) 3 and IRF7, promoting their translocation in the nucleus. To understand the molecular basis of activation of TBK1, we analyzed the sequence of TBK1 and IKK-i and identified a ubiquitin-like domain (ULD) adjacent to their kinase domains. Deletion or mutations of the ULD in TBK1 or IKK-i impaired activation of respective kinases, failed to induce IRF3 phosphorylation and nuclear localization and to activate IFN-beta or RANTES promoters. The importance of the ULD of TBK1 in LPS- or poly(I:C)-stimulated IFN-beta production was demonstrated by reconstitution experiments in TBK1-IKK-i-deficient cells. We propose that the ULD is a regulatory component of the TBK1/IKK-i kinases involved in the control of the kinase activation, substrate presentation and downstream signaling pathways.
Figures




Similar articles
-
Regulation and function of IKK and IKK-related kinases.Sci STKE. 2006 Oct 17;2006(357):re13. doi: 10.1126/stke.3572006re13. Sci STKE. 2006. PMID: 17047224 Review.
-
Involvement of TBK1 and IKKepsilon in lipopolysaccharide-induced activation of the interferon response in primary human macrophages.Eur J Immunol. 2007 Feb;37(2):528-39. doi: 10.1002/eji.200636090. Eur J Immunol. 2007. PMID: 17236232
-
Differential regulation of IKK alpha-mediated activation of IRF3/7 by NIK.Mol Immunol. 2008 Apr;45(7):1926-34. doi: 10.1016/j.molimm.2007.10.034. Epub 2007 Dec 18. Mol Immunol. 2008. PMID: 18068231
-
IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts.Proc Natl Acad Sci U S A. 2004 Jan 6;101(1):233-8. doi: 10.1073/pnas.2237236100. Epub 2003 Dec 16. Proc Natl Acad Sci U S A. 2004. PMID: 14679297 Free PMC article.
-
Interferon response induced by Toll-like receptor signaling.J Endotoxin Res. 2004;10(4):252-6. doi: 10.1179/096805104225005896. J Endotoxin Res. 2004. PMID: 15373970 Review.
Cited by
-
Crystal structure of inhibitor of κB kinase β.Nature. 2011 Apr 21;472(7343):325-30. doi: 10.1038/nature09853. Epub 2011 Mar 20. Nature. 2011. PMID: 21423167 Free PMC article.
-
Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates.J Cell Sci. 2013 Jan 15;126(Pt 2):580-92. doi: 10.1242/jcs.114926. Epub 2012 Nov 23. J Cell Sci. 2013. PMID: 23178947 Free PMC article.
-
Differential responses of normal human melanocytes to intra- and extracellular dsRNA.DNA Cell Biol. 2015 Jun;34(6):391-9. doi: 10.1089/dna.2014.2711. Epub 2015 Mar 24. DNA Cell Biol. 2015. PMID: 25803620 Free PMC article.
-
IKKε isoform switching governs the immune response against EV71 infection.Commun Biol. 2021 Jun 2;4(1):663. doi: 10.1038/s42003-021-02187-x. Commun Biol. 2021. PMID: 34079066 Free PMC article.
-
IκB kinase ε (IKKε): a therapeutic target in inflammation and cancer.Biochem Pharmacol. 2013 Apr 1;85(7):873-80. doi: 10.1016/j.bcp.2013.01.007. Epub 2013 Jan 17. Biochem Pharmacol. 2013. PMID: 23333767 Free PMC article. Review.
References
-
- Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124: 783–801 - PubMed
-
- Bayliss R, Sardon T, Vernos I, Conti E (2003) Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell 12: 851–862 - PubMed
-
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I (2005) Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 - PubMed
-
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

