Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample
- PMID: 17591929
- PMCID: PMC1899428
- DOI: 10.2353/jmoldx.2007.060170
Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample
Abstract
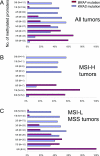
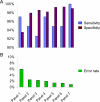
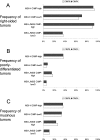
The CpG island methylator phenotype (CIMP or CIMP-high) with extensive promoter methylation is a distinct phenotype in colorectal cancer. However, a choice of markers for CIMP has been controversial. A recent extensive investigation has selected five methylation markers (CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1) as surrogate markers for epigenomic aberrations in tumor. The use of these markers as a CIMP-specific panel needs to be validated by an independent, large dataset. Using MethyLight assays on 920 colorectal cancers from two large prospective cohort studies, we quantified DNA methylation in eight CIMP-specific markers [the above five plus CDKN2A (p16), CRABP1, and MLH1]. A CIMP-high cutoff was set at > or = 6/8 or > or = 5/8 methylated promoters, based on tumor distribution and BRAF/KRAS mutation frequencies. All but two very specific markers [MLH1 (98% specific) and SOCS1 (93% specific)] demonstrated > or = 85% sensitivity and > or = 80% specificity, indicating overall good concordance in methylation patterns and good performance of these markers. Based on sensitivity, specificity, and false positives and negatives, the eight markers were ranked in order as: RUNX3, CACNA1G, IGF2, MLH1, NEUROG1, CRABP1, SOCS1, and CDKN2A. In conclusion, a panel of markers including at least RUNX3, CACNA1G, IGF2, and MLH1 can serve as a sensitive and specific marker panel for CIMP-high.
Figures
Comment in
-
The CpG island methylator phenotype in colorectal cancer.J Mol Diagn. 2007 Jul;9(3):281-3. doi: 10.2353/jmoldx.2007.070031. J Mol Diagn. 2007. PMID: 17591925 Free PMC article. No abstract available.
Similar articles
-
CpG island methylator phenotype-low (CIMP-low) colorectal cancer shows not only few methylated CIMP-high-specific CpG islands, but also low-level methylation at individual loci.Mod Pathol. 2008 Mar;21(3):245-55. doi: 10.1038/modpathol.3800982. Epub 2008 Jan 18. Mod Pathol. 2008. PMID: 18204436 Clinical Trial.
-
TGFBR2 mutation is correlated with CpG island methylator phenotype in microsatellite instability-high colorectal cancer.Hum Pathol. 2007 Apr;38(4):614-20. doi: 10.1016/j.humpath.2006.10.005. Epub 2007 Jan 31. Hum Pathol. 2007. PMID: 17270239
-
Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample.PLoS One. 2008;3(11):e3698. doi: 10.1371/journal.pone.0003698. Epub 2008 Nov 12. PLoS One. 2008. PMID: 19002263 Free PMC article.
-
Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review.Int J Cancer. 2011 Mar 1;128(5):1080-94. doi: 10.1002/ijc.25432. Int J Cancer. 2011. PMID: 20473920 Free PMC article. Review.
-
Different definitions of CpG island methylator phenotype and outcomes of colorectal cancer: a systematic review.Clin Epigenetics. 2016 Mar 2;8:25. doi: 10.1186/s13148-016-0191-8. eCollection 2016. Clin Epigenetics. 2016. PMID: 26941852 Free PMC article. Review.
Cited by
-
Stratification and Prognostic Relevance of Jass's Molecular Classification of Colorectal Cancer.Front Oncol. 2012 Feb 27;2:7. doi: 10.3389/fonc.2012.00007. eCollection 2012. Front Oncol. 2012. PMID: 22655257 Free PMC article.
-
Identification of a common variant with potential pleiotropic effect on risk of inflammatory bowel disease and colorectal cancer.Carcinogenesis. 2015 Sep;36(9):999-1007. doi: 10.1093/carcin/bgv086. Epub 2015 Jun 12. Carcinogenesis. 2015. PMID: 26071399 Free PMC article.
-
The Prognostic Value of Circulating Cell-Free DNA in Colorectal Cancer: A Meta-Analysis.J Cancer. 2016 Jun 4;7(9):1105-13. doi: 10.7150/jca.14801. eCollection 2016. J Cancer. 2016. PMID: 27326254 Free PMC article.
-
A prognostic CpG score derived from epigenome-wide profiling of tumor tissue was independently associated with colorectal cancer survival.Clin Epigenetics. 2019 Jul 24;11(1):109. doi: 10.1186/s13148-019-0703-4. Clin Epigenetics. 2019. PMID: 31340858 Free PMC article.
-
DNA methylation changes in ex-adenoma carcinoma of the large intestine.Virchows Arch. 2010 Oct;457(4):433-41. doi: 10.1007/s00428-010-0958-9. Epub 2010 Aug 14. Virchows Arch. 2010. PMID: 20711609
References
-
- Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14(Spec No 1):R65–R76. - PubMed
-
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. - PubMed
-
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

