RNAstrand: reading direction of structured RNAs in multiple sequence alignments
- PMID: 17540014
- PMCID: PMC1892782
- DOI: 10.1186/1748-7188-2-6
RNAstrand: reading direction of structured RNAs in multiple sequence alignments
Abstract
Motivation: Genome-wide screens for structured ncRNA genes in mammals, urochordates, and nematodes have predicted thousands of putative ncRNA genes and other structured RNA motifs. A prerequisite for their functional annotation is to determine the reading direction with high precision.
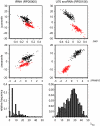
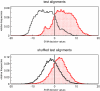
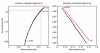
Results: While folding energies of an RNA and its reverse complement are similar, the differences are sufficient at least in conjunction with substitution patterns to discriminate between structured RNAs and their complements. We present here a support vector machine that reliably classifies the reading direction of a structured RNA from a multiple sequence alignment and provides a considerable improvement in classification accuracy over previous approaches.
Software: RNAstrand is freely available as a stand-alone tool from http://www.bioinf.uni-leipzig.de/Software/RNAstrand and is also included in the latest release of RNAz, a part of the Vienna RNA Package.
Figures
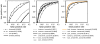

Similar articles
-
Hairpins in a Haystack: recognizing microRNA precursors in comparative genomics data.Bioinformatics. 2006 Jul 15;22(14):e197-202. doi: 10.1093/bioinformatics/btl257. Bioinformatics. 2006. PMID: 16873472
-
NcDNAlign: plausible multiple alignments of non-protein-coding genomic sequences.Genomics. 2008 Jul;92(1):65-74. doi: 10.1016/j.ygeno.2008.04.003. Epub 2008 Jun 3. Genomics. 2008. PMID: 18511233
-
LocARNA-P: accurate boundary prediction and improved detection of structural RNAs.RNA. 2012 May;18(5):900-14. doi: 10.1261/rna.029041.111. Epub 2012 Mar 26. RNA. 2012. PMID: 22450757 Free PMC article.
-
Sparse RNA folding revisited: space-efficient minimum free energy structure prediction.Algorithms Mol Biol. 2016 Apr 23;11:7. doi: 10.1186/s13015-016-0071-y. eCollection 2016. Algorithms Mol Biol. 2016. PMID: 27110275 Free PMC article. Review.
-
De novo discovery of structured ncRNA motifs in genomic sequences.Methods Mol Biol. 2014;1097:303-18. doi: 10.1007/978-1-62703-709-9_15. Methods Mol Biol. 2014. PMID: 24639166 Review.
Cited by
-
miRRim: a novel system to find conserved miRNAs with high sensitivity and specificity.RNA. 2007 Dec;13(12):2081-90. doi: 10.1261/rna.655107. Epub 2007 Oct 24. RNA. 2007. PMID: 17959929 Free PMC article.
-
GraphClust2: Annotation and discovery of structured RNAs with scalable and accessible integrative clustering.Gigascience. 2019 Dec 1;8(12):giz150. doi: 10.1093/gigascience/giz150. Gigascience. 2019. PMID: 31808801 Free PMC article.
-
Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA.BMC Genomics. 2013 Aug 9;14:543. doi: 10.1186/1471-2164-14-543. BMC Genomics. 2013. PMID: 23937650 Free PMC article.
-
Genome-Wide Analyses of MicroRNA Profiling in Interleukin-27 Treated Monocyte-Derived Human Dendritic Cells Using Deep Sequencing: A Pilot Study.Int J Mol Sci. 2017 Apr 28;18(5):925. doi: 10.3390/ijms18050925. Int J Mol Sci. 2017. PMID: 28452924 Free PMC article.
-
ViennaRNA Package 2.0.Algorithms Mol Biol. 2011 Nov 24;6:26. doi: 10.1186/1748-7188-6-26. Algorithms Mol Biol. 2011. PMID: 22115189 Free PMC article.
References
-
- Washietl S, Pedersen JS, Korbel JO, Gruber A, Hackermüller J, Hertel J, Lindemeyer M, Reiche K, Stocsits C, Tanzer A, Ucla C, Wyss C, Antonarakis SE, Denoeud F, Lagarde J, Drenkow J, Kapranov P, Gingeras TR, Guigó R, Snyder M, Gerstein MB, Reymond A, Hofacker IL, Stadler PF. Structured RNAs in the ENCODE Selected Regions of the Human Genome. Gen Res. 2007. - PMC - PubMed
LinkOut - more resources
Full Text Sources

