Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi's sarcoma-associated herpesvirus viral IRF homolog vIRF3
- PMID: 17522209
- PMCID: PMC1951281
- DOI: 10.1128/JVI.00235-07
Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi's sarcoma-associated herpesvirus viral IRF homolog vIRF3
Abstract
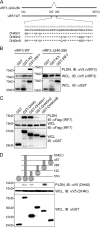
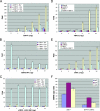
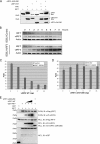
Upon viral infection, the major defense mounted by the host immune system is activation of the interferon (IFN)-mediated antiviral pathway that is mediated by IFN regulatory factors (IRFs). In order to complete their life cycle, viruses must modulate the host IFN-mediated immune response. Kaposi's sarcoma-associated herpesvirus (KSHV), a human tumor-inducing herpesvirus, has developed a unique mechanism for antagonizing cellular IFN-mediated antiviral activity by incorporating viral homologs of the cellular IRFs, called vIRFs. Here, we report a novel immune evasion mechanism of KSHV vIRF3 to block cellular IRF7-mediated innate immunity in response to viral infection. KSHV vIRF3 specifically interacts with either the DNA binding domain or the central IRF association domain of IRF7, and this interaction leads to the inhibition of IRF7 DNA binding activity and, therefore, suppression of alpha interferon (IFN-alpha) production and IFN-mediated immunity. Remarkably, the central 40 amino acids of vIRF3, containing the double alpha helix motifs, are sufficient not only for binding to IRF7, but also for inhibiting IRF7 DNA binding activity. Consequently, the expression of the double alpha helix motif-containing peptide effectively suppresses IRF7-mediated IFN-alpha production. This demonstrates a remarkably efficient means of viral avoidance of host antiviral activity.
Figures

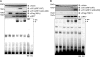

Similar articles
-
Duck Enteritis Virus Protein Kinase US3 Inhibits DNA Sensing Signaling by Phosphorylating Interferon Regulatory Factor 7.Microbiol Spectr. 2022 Dec 21;10(6):e0229922. doi: 10.1128/spectrum.02299-22. Epub 2022 Oct 26. Microbiol Spectr. 2022. PMID: 36287016 Free PMC article.
-
Downregulation of gamma interferon receptor 1 by Kaposi's sarcoma-associated herpesvirus K3 and K5.J Virol. 2007 Mar;81(5):2117-27. doi: 10.1128/JVI.01961-06. Epub 2006 Dec 13. J Virol. 2007. PMID: 17166914 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
XAF1 prevents hyperproduction of type I interferon upon viral infection by targeting IRF7.EMBO Rep. 2023 Jan 9;24(1):e55387. doi: 10.15252/embr.202255387. Epub 2022 Nov 17. EMBO Rep. 2023. PMID: 36394357 Free PMC article.
-
Pharmacological treatments in panic disorder in adults: a network meta-analysis.Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012729. doi: 10.1002/14651858.CD012729.pub3. Cochrane Database Syst Rev. 2023. PMID: 38014714 Free PMC article. Review.
Cited by
-
Functional Interfaces, Biological Pathways, and Regulations of Interferon-Related DNA Damage Resistance Signature (IRDS) Genes.Biomolecules. 2021 Apr 22;11(5):622. doi: 10.3390/biom11050622. Biomolecules. 2021. PMID: 33922087 Free PMC article. Review.
-
A Rhesus Rhadinovirus Viral Interferon (IFN) Regulatory Factor Is Virion Associated and Inhibits the Early IFN Antiviral Response.J Virol. 2015 Aug;89(15):7707-21. doi: 10.1128/JVI.01175-15. Epub 2015 May 13. J Virol. 2015. PMID: 25972548 Free PMC article.
-
Interplay between Kaposi's sarcoma-associated herpesvirus and the innate immune system.Cytokine Growth Factor Rev. 2014 Oct;25(5):597-609. doi: 10.1016/j.cytogfr.2014.06.001. Epub 2014 Jun 21. Cytokine Growth Factor Rev. 2014. PMID: 25037686 Free PMC article. Review.
-
Lysine 63-linked TANK-binding kinase 1 ubiquitination by mindbomb E3 ubiquitin protein ligase 2 is mediated by the mitochondrial antiviral signaling protein.J Virol. 2014 Nov;88(21):12765-76. doi: 10.1128/JVI.02037-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142606 Free PMC article.
-
Genome-Wide Transcriptional Roles of KSHV Viral Interferon Regulatory Factors in Oral Epithelial Cells.Viruses. 2024 May 25;16(6):846. doi: 10.3390/v16060846. Viruses. 2024. PMID: 38932139 Free PMC article.
References
-
- Au, W. C., Y. Su, N. B. Raj, and P. M. Pitha. 1993. Virus-mediated induction of interferon A gene requires cooperation between multiple binding factors in the interferon alpha promoter region. J. Biol. Chem. 268:24032-24040. - PubMed
-
- Bardos, J. I., and M. Ashcroft. 2005. Negative and positive regulation of HIF-1: a complex network. Biochim. Biophys. Acta 1755:107-120. - PubMed
-
- Baxevanis, A. D., and C. R. Vinson. 1993. Interactions of coiled coils in transcription factors: where is the specificity? Curr. Opin. Genet. Dev. 3:278-285. - PubMed
-
- Cassady, K. A., M. Gross, and B. Roizman. 1998. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620-8626. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

