Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers
- PMID: 17515920
- PMCID: PMC2714968
- DOI: 10.1038/nature05886
Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers
Abstract
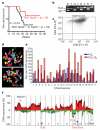
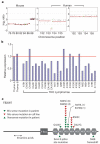
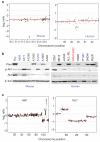
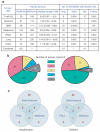
Highly rearranged and mutated cancer genomes present major challenges in the identification of pathogenetic events driving the neoplastic transformation process. Here we engineered lymphoma-prone mice with chromosomal instability to assess the usefulness of mouse models in cancer gene discovery and the extent of cross-species overlap in cancer-associated copy number aberrations. Along with targeted re-sequencing, our comparative oncogenomic studies identified FBXW7 and PTEN to be commonly deleted both in murine lymphomas and in human T-cell acute lymphoblastic leukaemia/lymphoma (T-ALL). The murine cancers acquire widespread recurrent amplifications and deletions targeting loci syntenic to those not only in human T-ALL but also in diverse human haematopoietic, mesenchymal and epithelial tumours. These results indicate that murine and human tumours experience common biological processes driven by orthologous genetic events in their malignant evolution. The highly concordant nature of genomic events encourages the use of genomically unstable murine cancer models in the discovery of biological driver events in the human oncogenome.
Figures
Similar articles
-
Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies.Genome Biol. 2016 May 31;17(1):115. doi: 10.1186/s13059-016-0971-7. Genome Biol. 2016. PMID: 27246460 Free PMC article.
-
Cooperative genetic defects in TLX3 rearranged pediatric T-ALL.Leukemia. 2008 Apr;22(4):762-70. doi: 10.1038/sj.leu.2405082. Epub 2008 Jan 10. Leukemia. 2008. PMID: 18185524
-
Combined MYC Activation and Pten Loss Are Sufficient to Create Genomic Instability and Lethal Metastatic Prostate Cancer.Cancer Res. 2016 Jan 15;76(2):283-92. doi: 10.1158/0008-5472.CAN-14-3280. Epub 2015 Nov 10. Cancer Res. 2016. PMID: 26554830 Free PMC article.
-
[Analysis of genomic copy number alterations of malignant lymphomas and its application for diagnosis].Gan To Kagaku Ryoho. 2007 Jul;34(7):975-82. Gan To Kagaku Ryoho. 2007. PMID: 17637530 Review. Japanese.
-
Pediatric leukemia/lymphoma with t(8;14)(q24;q11).Leukemia. 1992 Jul;6(7):613-8. Leukemia. 1992. PMID: 1385638 Review.
Cited by
-
Development of a colon cancer GEMM-derived orthotopic transplant model for drug discovery and validation.Clin Cancer Res. 2013 Jun 1;19(11):2929-40. doi: 10.1158/1078-0432.CCR-12-2307. Epub 2013 Feb 12. Clin Cancer Res. 2013. PMID: 23403635 Free PMC article.
-
Genome maintenance in the context of 4D chromatin condensation.Cell Mol Life Sci. 2016 Aug;73(16):3137-50. doi: 10.1007/s00018-016-2221-2. Epub 2016 Apr 20. Cell Mol Life Sci. 2016. PMID: 27098512 Free PMC article. Review.
-
Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice.Cell. 2007 Sep 7;130(5):863-77. doi: 10.1016/j.cell.2007.08.029. Cell. 2007. PMID: 17803909 Free PMC article.
-
MicroRNA-223 regulates cyclin E activity by modulating expression of F-box and WD-40 domain protein 7.J Biol Chem. 2010 Nov 5;285(45):34439-46. doi: 10.1074/jbc.M110.152306. Epub 2010 Sep 7. J Biol Chem. 2010. PMID: 20826802 Free PMC article.
-
Derivation of thymic lymphoma T-cell lines from Atm(-/-) and p53(-/-) mice.J Vis Exp. 2011 Apr 3;(50):2598. doi: 10.3791/2598. J Vis Exp. 2011. PMID: 21490582 Free PMC article.
References
-
- Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nature Genet. 2005;37(Suppl):S11–S17. - PubMed
-
- Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. - PubMed
-
- Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. - PubMed
-
- Sweet-Cordero A, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nature Genet. 2005;37:48–55. - PubMed
-
- Hodgson G, et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nature Genet. 2001;29:459–464. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

